AMD Activities: 2019
Posted on Tuesday, December 15, 2020
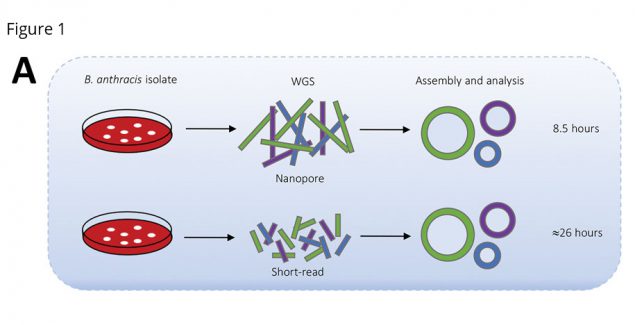
Anthrax is a rare but serious disease, requiring rapid identification to reduce illness and death. Bacillus anthracis–the bacterium that causes anthrax–is found naturally worldwide but has also been used as a biological weapon. Genetic characterization of B. anthracis can help health officials detect whether the samples are resistant to antibiotics or whether they have been genetically modified, information that can inform treatment and be useful in tracing the origin of the infection.
On August 2, 2019, CDC received a B. anthracis isolate (Ba0914) from an infected person in Texas who had been exposed to an animal that had died of anthrax. Conventional DNA sequencing takes 26 hours. Conventional antimicrobial susceptibility testing takes over 16 hours. But scientists in CDC’s Division of Preparedness and Emerging Infections used
a new method and a portable, pocket-sized nanapore sequencer. They were able to sequence the whole genome and confirm that the bacteria did not carry genetic markers for antibiotic resistance or evidence of genetic engineering in only 8.5 hours.
This study demonstrates the speed and usefulness of a portable sequencer. Although the same-day DNA sequencing results are not as precise as second-generation short-read sequencing, CDC had actionable data in approximately 1/3 the time. This can give public health decisionmakers the information they need to make rapid decisions during an emergency, when every minute counts.
Posted on Tuesday, December 15, 2020

In recent years, CDC has joined with other public health and regulatory officials to investigate troubling multistate outbreaks of E. coli infections linked to leafy greens. In one 2019 outbreak, 167 people in 27 states were infected with a strain of E. coli O157:H7. Whole-genome sequencing (WGS) showed that the bacteria from that outbreak were similar to the strain of E. coli O157:H7 from an outbreak linked to romaine lettuce a year earlier, in the fall of 2018, as well as a strain linked to leafy greens in late 2017. WGS data provided an early clue to investigators to consider leafy greens as a possible source of the 2019 outbreak. WGS revealed that samples taken from environments where lettuce was grown during some of these investigations also contained the same harmful strain of E. coli O157:H7 that was making people ill. These data led growers to adopt more stringent irrigation water standards to help reduce the risk of contamination in the future.
When combined with interview data from sick people and product traceback, the use of WGS data strengthens the U.S. food safety system by helping identify food vehicles and by providing information about how harmful bacteria like E.coli O157:H7 can contaminate food where it is grown. Since 2013, when PulseNet first began applying WGS in foodborne bacterial investigations, PulseNet and AMD have helped build national WGS capacity to detect and solve foodborne outbreaks with more precision than ever before. Lessons learned from this and other outbreak investigations will help guide industry and support government efforts to prevent similar occurrences in the future.
Posted on Tuesday, December 15, 2020
Infection with Helicobacter pylori, a digestive bacterium, has been linked to gastric cancer and ulcers and is common among Alaska Native people and people living in rural communities in Alaska. H. pylori can colonize and damage the protective lining of the stomach and establish infections that last for years or decades. To better understand this pathogen, scientists want to study its genetic data to look for markers related to H. pylori’s virulence and learn why it causes disproportionately high rates of gastric cancer in Alaska Native people.
CDC’s Arctic Investigations Program (AIP) leads epidemiology and laboratory efforts focused on H. pylori. With AMD support, CDC’s Division of Scientific Resources deployed a bioinformatician to work with AIP in Alaska for 6 months. By being on site and working directly with the AIP scientists, the bioinformatician was better able to create a Helicobacter pylori pipeline (HPype) to process, clean, and analyze H. pylori sequencing data. The bioinformatician modified existing resources to meet specific needs rather than creating something from scratch and established a computer system
to track and manage the data. Once all of the HPype analysis modules are complete, AIP will install them on the AMD portal to provide an interface for laboratorians and epidemiologists outside of Alaska so that they, too, can analyze the sequence data and learn more about this infectious disease.
Posted on Tuesday, December 15, 2020

CDC’s Influenza Division developed a rapid and reliable viral genomic sequencing kit called the Mobile Influenza Analysis (MIA). MIA is a portable influenza (flu) laboratory that can determine which flu strain is circulating on-site. The kit produces real-time data that can be used to evaluate whether a virus is new, what the associated public health risk is, and whether influenza vaccines against the circulating strain are available. These are key components to detecting and responding to new influenza strains and containing outbreaks before they start.
Posted on Thursday, December 26, 2019
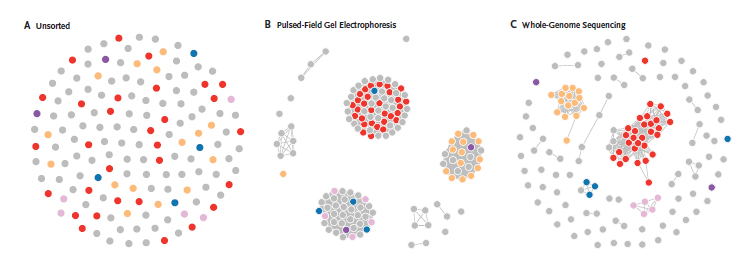
Each dot in these figures represent surveillance data for a foodborne pathogen, Salmonella enterica serovar Enteritidis. Panel A displays cases randomly, without regard to molecular subtyping. Panel B represents cases grouped by pulsed-field gel Electrophoresis (PFGE). The groupings include many cases unrelated to the outbreak, which complicates the investigation and reduces the likelihood of finding the food source. Panel C shows the finer resolution afforded by whole-genome sequencing was more effective in segregating outbreak cases.
A December 26 report by CDC scientists and colleagues in the New England Journal of Medicine (NEJM) highlights how a set of new technologies, collectively known as “next-generation sequencing,” have revolutionized scientists’ ability to decode DNA. These technologies are transforming the response to infectious disease outbreaks, providing new insights into disease emergence and transmission, expediting pathogen characterization, and promoting data sharing.
The AMD program has shown that CDC and other public health scientists can quickly and effectively adapt, incorporate and deploy novel, highly effective and transformative technologies into the public health system. Before AMD, the U.S. public health system (both CDC and state and local health departments) was lagging in the use of innovative new laboratory technologies. Now, it is a global leader. CDC and state and local public health agencies are now using these technologies on a daily basis to detect outbreaks quicker, investigate them more effectively, get a clearer understanding of disease transmission and, ultimately, develop better approaches to prevent infections and outbreaks of infectious disease from occurring in the first place.
You can read more about the transformative role pathogen genomics have on public health in the December 26th special report from the NEJM.
Posted on Wednesday, November 13th, 2019

Next-generation Sequencing (NGS) is evolving quickly, and even though CDC, APHL, and others have published guidance documents, a need persists for additional laboratory resources.
The NGS Quality Initiative is developing an NGS-focused quality management system (QMS) to address the many challenges public health and clinical laboratories encounter when they develop and implement NGS-based tests, by providing ready-to-implement guidance documents, customizable standard operating procedures, and other tools. The project is funded by CDC’s Office of Advanced Molecular Detection, and is co-led by the Division of Laboratory Systems, the Office of the Deputy Director for Infectious Diseases, and the Association of Public Health Laboratories.
Posted on Thursday, October 10th, 2019
In August 2016, a community of homeless people in Boston started showing up at a clinic with HIV. By June 2018, the outbreak involved more than 120 people, predominantly in the cities of Lawrence and Lowell in northeastern Massachusetts. The Massachusetts Department of Public Health (MDPH) asked for CDC’s help in investigating and responding to the outbreak.
Posted on Wednesday, August 21th, 2019
Ah, summertime at the beach; a time to relish the sun, the sand, and the ocean. Millions of people flock to the seashore each year to enjoy summertime fun. One resort town in New Hampshire draws many visitors from neighboring states each summer to its sandy beaches along the Atlantic shore. But in the summer of 2018, with the town’s annual seafood festival just weeks away, the town faced an unwelcomed visitor. Several people landed in the hospital with a severe form of pneumonia after visiting the town. A team of state, local, and federal investigators leapt into action to find the cause of the illnesses.
Posted on Friday, May 24th, 2019
Recent research in the Streptococcus Lab (StrepLab) at CDC is improving how scientists identify outbreaks of group A strep infections. While CDC’s scientists were developing the cluster detection tool in 2017, epidemiologists in Minnesota noticed an increase in these kinds of infections. StrepLab’s new technology helped researchers detect and investigate a deadly multi-facility outbreak that might have otherwise been missed.
Posted on Friday, May 24th, 2019

This year, the PulseNet national laboratory network is transitioning from pulsed-field gel electrophoresis (PFGE) to whole genome sequencing (WGS) to combat foodborne diseases more effectively. PulseNet public health scientists from laboratories across the United States will now have the tools to generate, analyze, and share their WGS results of foodborne bacteria with the other network participants.
Posted on Monday, May 13th, 2019
Cyclospora cayetanensis is a particularly challenging pathogen to analyze, but thanks to AMD and a prototype typing tool that creates a “DNA fingerprint,” it may become easier for disease detectives to crack the code on Cyclospora outbreaks. CDC scientists are prepared to type samples in real time during the 2019 Cyclospora season to evaluate the new tool’s utility for outbreak detection. With this new tool, researchers hope to assist public health officials to detect more outbreaks and to learn how these pathogens are getting into the food system.

Posted on Monday, April 8, 2019
The AMD program has partnered with the CDC Foundation and the Bill and Melinda Gates Foundation (BMGF) on a one-year project to define what software infrastructure is needed to support the complex information workflows required by AMD technologies.
In March, OAMD’s Greg Armstrong and Duncan MacCannell and NCIRD’s Elizabeth Neuhaus met with the project team, which also includes Africa CDC officials, Trevor Bedford’s laboratory at the University of Washington, and a group of experts in bioinformatics and open software development. The group seeks consensus on a sustainable framework for this software—a framework that will facilitate the application of pathogen genomics to public health both in high-income countries such as the United States as well as in low- and middle-income countries.
AMD Director Greg Armstrong said, “It was remarkable how strongly the participants felt about the need for this meeting and for an effort to coordinate the development of software that will support pathogen genomics in public health.”
Posted on Monday, March 11, 2019
The Office of Advanced Molecular Detection (OAMD) welcomed epidemiologists and microbiologists from state and local health departments across the nation to the first AMD Academy, hosted with the Association of Public Health Laboratories (APHL) and the Council of State and Territorial Epidemiologists (CSTE). Held in Atlanta during the last week of January, OAMD presented a 2-day molecular epidemiology course for epidemiologists (January 30–31) and a 4-day, intermediate-level bioinformatics course for microbiologists (January 28–31).
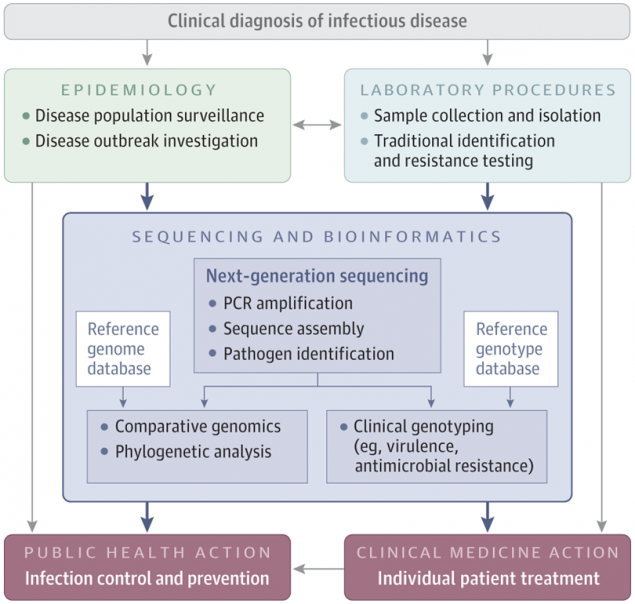
Posted on Tuesday, March 5, 2019
On February 14, the Journal of the American Medical Association (JAMA) published an article describing ongoing advances being made by CDC’s Advanced Molecular Detection (AMD) program in the field of next generation sequencing (NGS). JAMA also published a related clinical review podcast, where they interviewed two of the authors, Marta Gwinn, MD, MPH, and Gregory L. Armstrong, MD, from the Office of AMD about the potential NGS holds for improving clinical and public health microbiology.

