Genetics, coffee consumption, and Parkinson’s disease Questions
Parkinson’s disease (PD) is the second most common neurodegenerative disease after Alzheimer’s disease. Population prevalence of PD increases from about 1% at age 60 to 4% by age 80. Early symptoms of PD include tremor, rigidity, and difficulty walking; cognitive decline is common at later stages. The underlying pathology of PD is selective death of dopamine-generating cells in the substantia nigra, a part of the brain involved in movement, reward, and addiction. Treatment of PD with levodopa temporarily controls motor symptoms but does not slow disease progression. Like other common diseases, PD is thought to arise from complex interactions between genetic and environmental factors, which remain mostly unknown.
Many epidemiologic studies have found that PD is less common in people who drink coffee, as well as in those who smoke cigarettes. These findings are remarkably consistent and they have survived an exhaustive search for non-causal explanations. No one would suggest taking up smoking (or even coffee drinking) to prevent PD; however, understanding their “protective effects” could shed light on its molecular pathology. Such insights could suggest new treatments that not only treat symptoms but can slow or halt disease progression.
Question 1: Several case-control studies of PD have examined interactions of coffee consumption with candidate genes that were selected because of their associations with PD (e.g., SNCA), with caffeine metabolism (e.g., CYP1A2), or with caffeine’s CNS effects (e.g., ADORA2A). Although these studies have often confirmed previously observed main effects, they have not found statistically significant gene-environment interactions. Suggest two possible reasons why such studies have been unsuccessful in identifying interactions.

Question 2: From 2005 through mid-2012, 47 genome-wide association studies (GWAS) of PD reported 79 “hits” (highly significant associations) in 66 genes. Until 2011, none of these GWAS had examined any environmental risk factors (or protective factors). Comment on two challenges to studying gene-environment interactions in a GWAS.

The remaining questions are related to the following study, which evaluated a candidate exposure (coffee drinking) and searched across the genome for interactions, treating all SNPs equally regardless of prior association with PD, coffee drinking, or caffeine metabolism.
Hamza TH, et al. Genome-Wide Gene-Environment Study Identifies Glutamate Receptor Gene GRIN2A as a Parkinson’s Disease Modifier Gene via Interaction with Coffee. PLoS Genet 2011;7(8): e1002237.
Author Summary
Parkinson’s disease (PD), like most common disorders, involves interactions between genetic make-up and environmental exposures that are unique to each individual. Caffeinated-coffee consumption may protect some people from developing PD, although not all benefit equally. In a genome-wide search, we discovered that variations in the glutamate-receptor gene GRIN2A modulate the risk of developing PD in heavy coffee drinkers. The study was hypothesis-free, that is, we cast a net across the entire genome allowing statistical significance to point us to a genetic variant, regardless of whether it fell in a genomic desert or an important gene. Fortuitously, the most significant finding was in a well-known gene, GRIN2A, which regulates brain signals that control movement and behavior. Our finding is important for three reasons: First, it is a proof of concept that studying genes and environment on the whole-genome scale is feasible, and this approach can identify important genes that are missed when environmental exposures are ignored. Second, the knowledge of interaction between GRIN2A, which is involved in neurotransmission in the brain, and caffeine, which is an adenosine-A2A-receptor antagonist, will stimulate new research towards understanding the cause and progression of PD. Third, the results may lead to personalized prevention of and treatment for PD.
In the Methods section, the authors explained their choice of a joint test of SNP main effect and its interaction with coffee:
The main advantage of the joint test is that it does test for interaction and it has more power than pure interaction test when there is a modest SNP marginal effect.
The joint test was proposed by Kraft et al. in Exploiting gene-environment interaction to detect genetic associations (Hum Hered 2007;63:111-9). In that article, Kraft compared four approaches to testing for gene-environment interaction in case-control studies:
- Assume that a marginal genetic effect will also be detectable in the setting of gene-environment interaction, so test only for gene-disease association (G test);
- Test for main genetic and environmental effects and include an interaction term (GE test);
- Test for genetic association, allowing for heterogeneity of effect across exposure strata (G-GE test, or joint test);
- Screen markers for departures from multiplicative interaction, for example by using a case-only design (GEca test).
The data below (excerpted from Table 3 in Hamza et al.) summarize the effects of GRIN2A genotype and coffee intake (adjusted for sex and age at interview). P-values for the stratified analysis (genotype-coffee effects) are based on the joint test (G-GE):
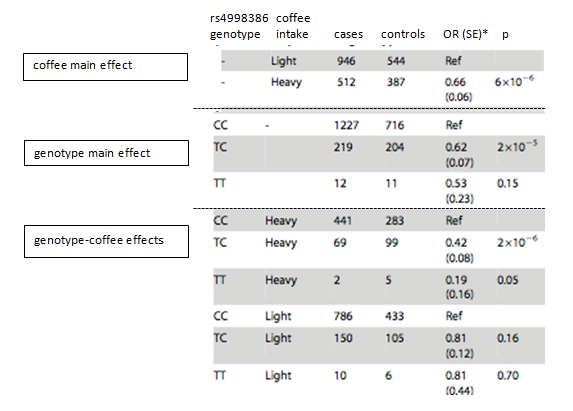
Question 3: Approximately how similar/different are the main effects of GRIN2A genotype and coffee intake?

Question 4: The authors stated that they tested models of “Dominant, Additive, and Recessive modes of inheritance” of the implicated GRIN2A allele. What does this statement mean?

Question 5:
| Genotype | Coffee | Cases | Controls | OR(calc) | 95% CI | p-value (chisq) |
|---|---|---|---|---|---|---|
| CC | Light | |||||
| CC | Heavy | |||||
| TC/TT | Light | |||||
| TC/TT | Heavy |

Question 6: Use the case data from your table in 5. to perform a case-only analysis. Note that because coffee intake and the T allele have protective effects against PD, all the OR’s will be <1.0. In the absence of gene-environment interaction, the “synergy index,” which is calculated as ORge / (ORg * Ore) is expected to equal 1.0. Calculate the synergy index.
Is ORge / ( ORg * ORe ) equal to 1.0?

| Heavy |
Light |
|
|---|---|---|
| TC/TT | ||
| CC |
Question 7: Is coffee intake independent of genotype?

Reviewing the findings of this study in terms of the four approaches to testing for gene-environment interaction described above:
- G test: The marginal effect for the TC genotype from the authors’ adjusted model was OR=0.62, p < 2×10-5 in the discovery study. The authors noted that this would not have been considered significant according to typical GWAS significance threshold.
- GE test: The authors did not present results for a “pure interaction test.” The results calculated for question 5 above would be a starting point for such an analysis.
- G-GE test: This was the basis for p-values reported in Table 3.
- GEca test: The case-only analysis requires independence of G and E.
Question 8: In the Abstract, the authors made three arguments for the importance of their findings. Based on your evaluation of the data presented in these questions (you are not required to review the entire article), how persuaded are you of each of their claims? Use the “smiling scale” to consider how you would rate each argument, then write 2 or 3 sentences to explain your ratings overall.
Example: This article makes me want to drink more coffee.
Argument 1
First, it is a proof of concept that studying genes and environment on the whole-genome scale is feasible, and this approach can identify important genes that are missed when environmental exposures are ignored.
Argument 2
Second, the knowledge of interaction between GRIN2A, which is involved in neurotransmission in the brain, and caffeine, which is an adenosine-A2A-receptor antagonist, will stimulate new research towards understanding the cause and progression of PD.
Argument 3
Third, the results may lead to personalized prevention of and treatment for PD.
