Chapter 22 – Evaluation of predictive genetic tests for common diseases: bridging epidemiological, clinical, and public health measures Tables
Human Genome Epidemiology (2nd ed.): Building the evidence for using genetic information to improve health and prevent disease
necessarily represent the views of the funding agency.”
A. Cecile J. W. Janssens, Marta Gwinn, and Muin J. Khoury
| Measure | Description | Formula |
|---|---|---|
| Epidemiological evaluation | ||
| Genotype frequency | Frequency of genotype in total population | P(g) = g/N |
| Population risk | Disease risk in total population | P(D) = D/N |
| Penetrance | Disease risk conditional on genotype status | 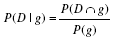 |
| Relative risk | Ratio of disease risks of carriers and noncarriers | RR = P(D | G) / P(D | n G) |
| Odds ratio | Ratio of odds of disease of carriers and noncarriers | 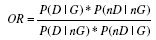 |
| Risk difference | Difference between disease risks of carriers and noncarriers | RD = P(D | G)−P(D | n G) |
| Clinical validity | ||
| Sensitivity | Proportion of carriers among affected | Se = P(G|D) |
| Specificity | Proportion of noncarriers among unaffected | Sp = P(nG|nD) |
| False positive rate | Proportion of carriers among unaffected | FPR = 1-Sp = P(G|nD) |
| False negative rate | Proportion of noncarriers among affected | FNR = 1-Se = P(nG|D) |
| Positive predictive value | Proportion of affected among carriers | PPV = P(D|G) |
| Negative predictive value | Proportion of unaffected among noncarriers | NPV = P(nD|nG) |
| Clinical or public health utility | ||
| Likelihood ratio | Ratio of the genotype frequency in affected and the genotype frequency in unaffected | LRg = P(g|D)/P(g|nD) |
| Population attributable fraction | Proportion of cases that is attributable to the genetic variant | 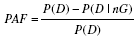 |
| Number needed to treat | Number needed to treat to prevent one case | 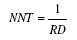 |
| Number needed to screen | Number of cases needed to screen to prevent one case | 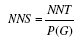 |
Persons with the risk genotype are called “carriers”; those without the risk genotype are “noncarriers.” Persons who will develop the disease are called “affected”; those who will not are called “unaffected.” The letter “g” represents the number of persons with a given genotype, which can be either the risk genotype (G) or the referent genotype (nG). N = the total number of persons in a population; D = the number of persons who will develop the disease; nD = the number of persons who will not develop the disease; P = probability. The symbol “|” stands for “conditional on”: for example, P(D|G) means “the probability of disease conditional on the risk genotype,” or “the proportion of persons with the risk genotype who will develop disease.” The symbol “∩” denotes “and.” Although the examples refer to predictive testing for future disease, all measures can also be calculated for diagnostic tests that aim to identify persons with or without the disease.
| Will develop disease | Will not develop disease | Total | |
|---|---|---|---|
| Carriers of risk genotype | True positive | False positive | G |
| Noncarriers | False negative | True negative | nG |
| Total | D | nD | N |
Persons with the risk genotype are called “carriers”; those without the risk genotype are “noncarriers.” The letter “g” represents the number of persons with a given genotype, which can be either the risk genotype (G) or the referent genotype (nG). N = the total number of individuals in a population; D = the number of individuals who develop the disease; nD = the number of individuals who will not develop the disease.
| Disease | Huntington disease | Breast cancer | Colorectal cancer | Type 2 diabetes | |||
|---|---|---|---|---|---|---|---|
| Clinical scenario* | Offspring of patients | Offspring of mutation carriers | General population | ||||
| Gene | 4p16.3 | BRCA1/2 | MLH1/MSH2 | PPARG | CAPN10 | TCF7L2 | |
| Marker | CAG repeats | P12A | SNP44 | rs7903147 | |||
| Genotype definition | At-risk | Mutations | Mutations | Mutations | PP | TT | TT |
| Referent | PA/AA | CC/CT | CC/CT | ||||
| Epidemiological evaluation | |||||||
| Genotype frequency | At-risk | 50% | 50% | 50% | 73% (18) | 62% (18) | 7% (13) |
| Referent | 50% | 50% | 50% | 26% | 38% | 93% | |
| Disease risk | 50% | 39%† | 38%† | 33% (14) | 33% (14) | 33% (14) | |
| Penetrance | At-risk | 100% | 65% (19) | 70% (9) | 36% | 36% | 49% |
| Referent | 0% | 13% (20) | 6% (20) | 24% | 28% | 32% | |
| Odds ratio | ∞ | 12.9 | 40.1 | 1.77 (18) | 1.45 (18) | 2.05 (13) | |
| Relative risk | ∞ | 5.13 | 12.6 | 1.49 | 1.29 | 1.54 | |
| Risk difference | 100% | 52% | 64% | 12% | 8% | 17% | |
| Clinical validity and utility | |||||||
| Sensitivity | 100% | 84% | 93% | 80% | 68% | 10% | |
| Specificity | 100% | 72% | 76% | 31% | 41% | 95% | |
| False negative rate | 0% | 29% | 24% | 70% | 59% | 90% | |
| False positive rate | 0% | 16% | 7% | 20% | 32% | 5% | |
| Positive predictive value | 100% | 65% | 70% | 36% | 36% | 49% | |
| Negative predictive value | 100% | 87% | 94% | 76% | 72% | 68% | |
| Likelihood ratio | At-risk | ∞ | 2.94 | 3.88 | 1.15 | 1.15 | 1.94 |
| Referent | 0 | 0.23 | 0.1 | 0.65 | 0.79 | 0.95 | |
| Clinical or public health impact | |||||||
| Population attributable fraction | 100% | 67% | 85% | 26% | 15% | 4% | |
| Number needed to treat | 1 | 2 | 2 | 8 | 12 | 6 | |
| Number needed to screen | 2 | 4 | 3 | 11 | 20 | 84 | |
Numbers are for illustration purposes only. Specific risk estimates may vary among populations. All calculations were performed according to the formulas from Table 22.2, by using the Risk Translator of the HuGE Navigator.
*Clinical scenario specifies the target population for the genetic testing.
†Disease risks for offspring of mutation carriers, calculated as the average of the penetrances.