Accelerating the collection and interpretation of data
Increased computing power to fight COVID-19
When exhausting all possibilities to fight a pandemic, it’s helpful to have virtually inexhaustible computational capacity, and the Advanced Molecular Detection (AMD) program’s Scientific Computing (SciComp) resource delivers flexible, powerful tools to CDC and public health departments for use against COVID-19.
Over the past 7 years, AMD has assembled a state-of-the-art computing center infrastructure (see below). The SciComp environment hosts systems and applications critical to the COVID-19 pandemic response. One system powers a dashboard that allows real-time analysis and visualization of syndromic surveillance data from across the nation. Another application, built by AMD SciComp developers, lets state public health labs get much clearer results with COVID-19 tests, while another eases shortages of COVID-19 test kits by matching unused supplies with places that need more kits. SciComp is modernizing data collection and analysis at CDC as big data continues to reshape health sciences.

SciComp infrastructure includes:
- 140+ computing machines filling 15 racks
- 750+ virtual computing machines
- 30+ specialized computing systems
Provides:
- 4+ petabytes of fast, efficient, on-premise storage to expedite COVID-19 genomic data analysis
- Cloud archiving for efficient off-site backup and archiving
Networks:
- 100+ lab instruments uploading data in real time to permit rapid analysis
- 2000+ user accounts support the expanding computational needs of the agency
Building a network to share lab test results years before COVID-19 struck
Prior to COVID-19, NCEZID made critical and strategic investments through the Epidemiology and Laboratory Capacity for Prevention and Control of Emerging Infectious Diseases (ELC) Cooperative Agreement to ensure that more than 90 percent of state and local health departments had widespread use of electronic laboratory reporting within their jurisdictions.
As early as 2005, CDC, in partnership with the Association of Public Health Laboratories, began paving the way for what is now the COVID-19 Electronic Laboratory Reporting (CELR) program. They created a secure, cloud-based platform to help state health departments, state laboratories, regional commercial laboratories, and federal agencies report their data. State health departments have increasingly used this platform, supported by ongoing financial and technical support from CDC, to increase the proportion of the reportable disease results they share with CDC.
Building upon these advances allowed the CELR program to quickly enable detailed data reporting from public health departments to CDC for approximately 90 percent of the COVID-19 testing volume conducted nationwide. This robust dataset, which has been stripped of personally identifiable information, informs public health decision making to help reduce disease in communities.
Following COVID-19’s genetic breadcrumbs
CDC’s Advanced Molecular Detection (AMD) program has built a national network of more than 600 scientists who are using genetic data to track COVID-19 and fight its spread. Representing more than 150 national laboratories, universities, medical companies, foundations, and health departments, these scientists have joined forces to form SARS-CoV-2 Sequencing for Public Health Emergency Response, Epidemiology and Surveillance (SPHERES).
The genetic blueprint, or genome, of SARS-CoV-2, the virus that causes COVID-19, changes gradually as the virus reproduces and spreads from person to person. Public health scientists track these changes to follow the disease’s path through the world like a trail of breadcrumbs. They then identify clusters of people who caught COVID-19 from the same sources. This helps public health professionals zero in on outbreak hotspots to find communities that need increased testing and treatment. Keeping tabs on genomic changes is needed to track new variants and to monitor the impact they could have on diagnostic tests, drugs, and vaccines. This requires a lot of samples, data, and coordination. The SPHERES consortium brings labs together to make this detective work possible. AMD’s role is to coordinate efforts to ensure that the data being generated across hundreds of laboratories are rapidly available and have the greatest impact.
Making data on foodborne diseases easier to digest
Because seeing is believing, CDC is using data visualization to help people better understand threats from foodborne diseases. CDC’s FoodNet Fast web tool allows users to effortlessly interact with color-coded graphs of nine common infectious microbes that grow in food. For example, you can see in the graph above how rates of illness caused by different foodborne pathogens have changed over the years. This graph shows that Salmonella and Campylobacter are the most common foodborne microbes. In 2020, FoodNet Fast was updated to show users how different types of diagnostic tests are impacting the overall number of reported cases of foodborne illness. This streamlined searchable database offers instantaneous search results to anyone curious about how rates of illness have changed since 1996.
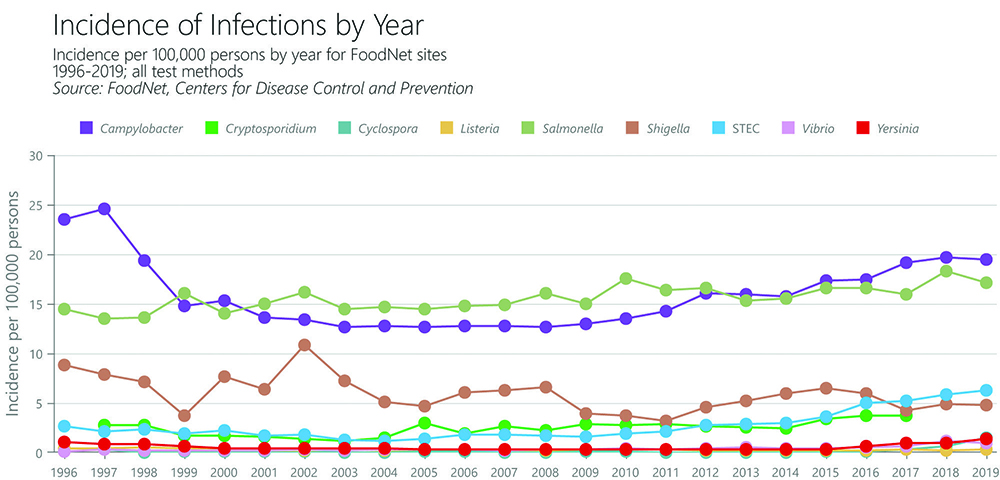
FoodNet Fast’s Pathogen Surveillance Tool shows how rates of illnesses have changed in FoodNet’s surveillance area since 1996 for selected pathogens transmitted commonly through food.

On accelerating genomic sequencing
“Sequence data sharing is an incredibly powerful tool in understanding this virus and in responding to the pandemic.”
Duncan MacCannell, AMD Chief Science Officer, NCEZID
