NCEZID Innovations: Whole Genome Sequencing
DNA Test Makes Food Safer
How quickly would you like to learn about an outbreak associated with foods you typically eat? The sooner the better. Rapid and precise identification of the bacteria causing foodborne illness is critical for timely foodborne outbreak response. Existing laboratory methods, used for many years to identify and describe bacterial foodborne pathogens, are complex and typically take 1 to 3 weeks to complete.
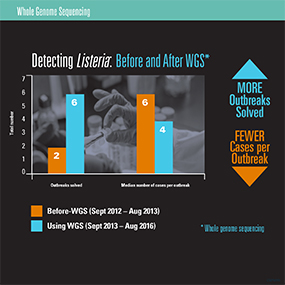
The good news is that a laboratory method called whole genome sequencing (WGS) is already transforming the way we identify and describe foodborne bacterial pathogens, and CDC is expanding its reach. WGS gives an exact DNA profile of bacteria that disease detectives can use to detect outbreaks and help identify a possible food culprit. WGS is an advanced molecular detection technology that provides scientists with more data in less time. This makes it possible to detect and solve foodborne outbreaks faster, resulting in fewer people getting sick. CDC’s PulseNet, a national laboratory network, is using WGS to improve its process for finding the most common foodborne pathogens, such as Listeria and Salmonella.
Now, NCEZID is making WGS testing available to all public health laboratories in the United States, providing a more complete national picture of foodborne illnesses than we have ever had. The goal is to convert PulseNet to a fully WGS-based network by the end of 2018. The lessons learned through WGS will help the industry to produce safer food and prevent future illnesses.
Download printable PDF version: Innovations to Stop Emerging and Zoonotic Infections [32 pages]
NCEZID foodborne disease scientists are using a new method to more quickly and accurately detect outbreaks.