Technical Updates

Patching Schedule
- November 15: NSSP vendor patches (Testing and Development) 12:00–8:00 AM ET
- November 17: NSSP vendor patches (Onboarding and Production) 12:00–8:00 AM ET
- December 20: NSSP vendor patches (Testing and Development) 12:00–8:00 AM ET
- December 22: NSSP vendor patches (Onboarding and Production) 12:00–8:00 AM ET
Jupyter Added to BioSense Platform RStudio Workbench
Effective October 26, 2022, NSSP added Jupyter Notebook and JupyterLab to the BioSense Platform RStudio Workbench.
- Jupyter Notebook is a web application for creating and sharing computational documents. Notebook is the classic notebook interface.
- JupyterLab is a web-based interactive development environment (IDE) for notebooks, code, and data. Its interface allows users to configure and arrange workflows. A modular design invites extensions to expand and enrich functionality. JupyterLab is a next generation IDE notebook interface.
NSSP notified licensed users of RStudio Workbench and provided access instructions. Licensed users who are not familiar with these products can review the documentation at Jupyter.org.
New Tab Added to ESSENCE Library
Given recent surges in Respiratory Syncytial Virus (RSV), we’ve added a MyESSENCE tab to the ESSENCE Library to help with surveillance. The tab is titled “RSV Surveillance—ED and Lab Combined (Full Details).” It integrates emergency department and commercial lab data, with 10-year, age-stratified trends for both data sources.
To access the tab: Log in to ESSENCE, click on the MyESSENCE tab, and then click Library. Search for RSV in the library search bar. The dashboard name should pop up. Select the check box and then click Download.
As you download and use the tab information, let us know how it works for you. It’s also easy to add additional widgets and stratifications, so help us make future improvements by contacting nssp@cdc.gov and making us aware of your preferences.
Onboarding Updates
A new Slack channel has been created. Join #mortality-data as a space to collaborate, ask questions, and share best practices and more for working with the mortality data source.
The NSSP onboarding team is available as a resource for staff who are helping with facility onboarding. New and refresher training can be requested for onboarding and validation by emailing nssp@cdc.gov or by submitting an NSSP Service Desk ticket.
How Sites Route Mortality Data to NSSP
The National Syndromic Surveillance Program (NSSP) has been refining its approach to routing mortality data to NSSP–ESSENCE using findings from a 2020 pilot project. The pilot activities have helped the NSSP team identify successful options for onboarding a site’s mortality data. However, standardizing the process to exchange mortality data can still be challenging due to variation in each site’s organizational structure. NSSP and its onboarding team have been working diligently to understand the complexities that exist within sites and to develop routing guidance to assist with mortality data onboarding.
The question is often asked, how are mortality data routed to NSSP’s BioSense Platform? A simplified routing diagram is included in the Mortality Engagement Packet. But for those site administrators and analysts who are not familiar with vital statistics and records offices, NSSP developed an overview diagram (Figure 1) to show mortality data routing.* Keep in mind that routing may vary from site to site and still produce high-quality data for surveillance and analyses.
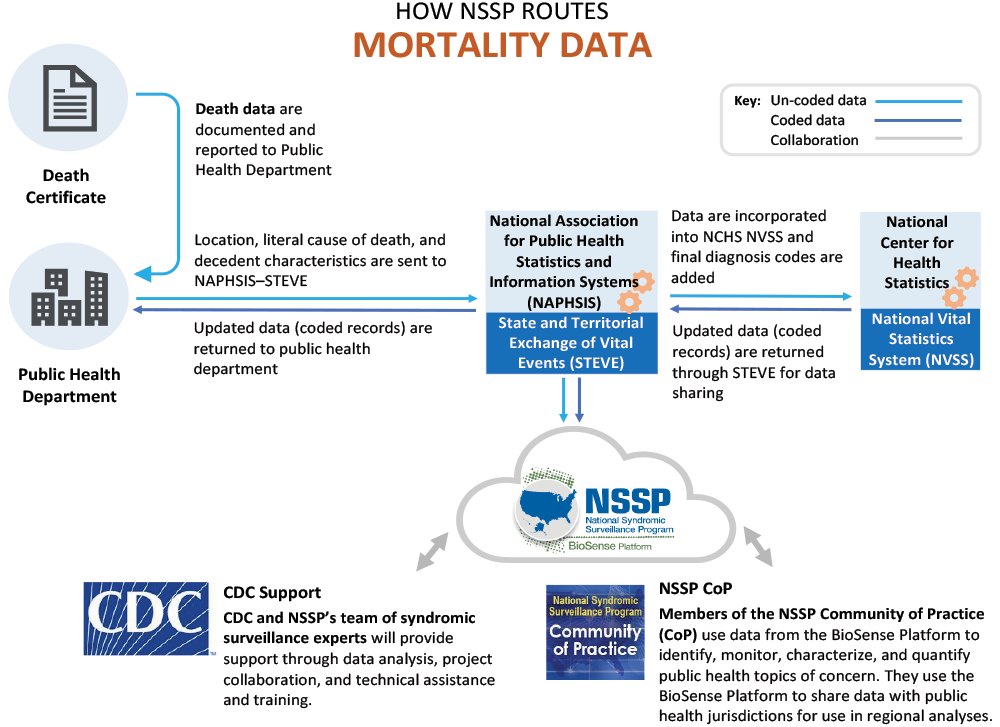
General Overview of Routing Death Information
Typically, the certifier (physician, coroner, medical examiner) declares the literal cause of death for the decedent and adds this information to the death certificate. Next, the death certificate is sent to the site’s public health department (or a site’s vital records office) where the literal cause of death information is routed (via State and Territorial Exchange of Vital Events, STEVE) to the National Center for Health Statistics (NCHS). NCHS updates the death records by assigning ICD-10 codes based on the literal cause of death. NCHS then routes the updated (coded information) back to the site using the same information exchange platform, STEVE.
The preferred file format is the standard Inter-Jurisdictional Exchange (IJE) electronic file format defined by the National Association for Public Health Statistics and Information Systems (NAPHSIS) IJE Committee.
How to Participate
Sites interested in incorporating their mortality data into NSSP may participate by contacting nssp@cdc.gov. Executing a data use agreement (DUA) is at the discretion of the site. NSSP does not require a DUA but can provide one. A standard NSSP DUA and template of the IJE file format will be shared upon request.
*The diagram shows the preferred flow for mortality data that was identified through the pilot.
Resources:
October NSSP Community of Practice Call
FAQs on Mortality Data Source
Slack channel: #Mortality-data NEW
If you liked this article, see NSSP’s Onboarding web page to read about moving data into production, why sites should onboard new facilities, and more.
Publication Updates
- Query Development: Community and Technical Resources—A time-saver for sure, this job aid pulls together all aspects of developing a query. It defines the process, explains how to create the query (syndrome definition) from searching ICD codes to consulting others via Slack. The job aid provides scores of resources (articles, guidance documents, and self checks) and instructions for revising queries. First timers who follow this guide will have much more confidence in their final product.
- Ultimate Guide for Site Administrators—This job aid highlights the community’s best practices. It suggests ways to connect with peers and exchange technical solutions quickly. It points to all things NSSP and is guaranteed to jump start your learning about NSSP–ESSENCE. Originally intended for site administrators, this job aid will also help new entrants to syndromic surveillance get acclimated quickly.
- An article has been added to the Onboarding webpage: “When to Use Associated Facilities: Guidance for Site Administrators.” Authored by the NSSP onboarding team, the article explains how associated facilities can function as a single entity so that ESSENCE can display data in a time-series graph.
Replication and the DataMart
NSSP replicates data to the Analytic DataMart so that analytics and validation don’t slow the processing of incoming data.
When syndromic data are received by the BioSense Platform, they are scrubbed to remove personally identifying information (PII), ingested, and then processed into the ESSENCE application for use in surveillance activities. To optimize processing, one database table is allocated to each site and data are indexed.
An overview of the data flow is described in the document Data Dictionary: Data Elements Used in NSSP Data Processing Journey. More information on data element processing and indexing is available in the NSSP Data Dictionary.
Shown below is a detailed view of how data are processed within the Archive database and replicated to the Analytic DataMart (Figure 1) where the archiving process fits into the larger scheme of data processing (Figure 2).
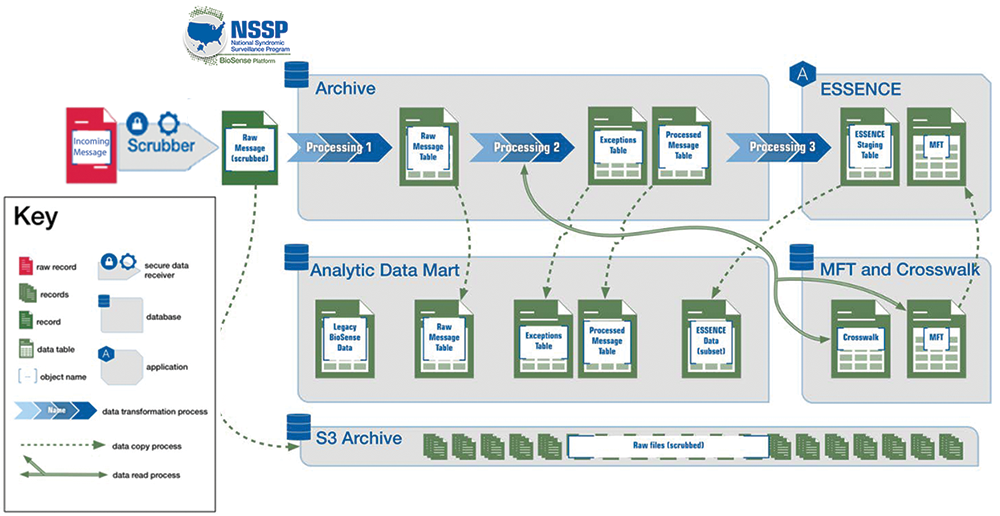
Detailed view below:
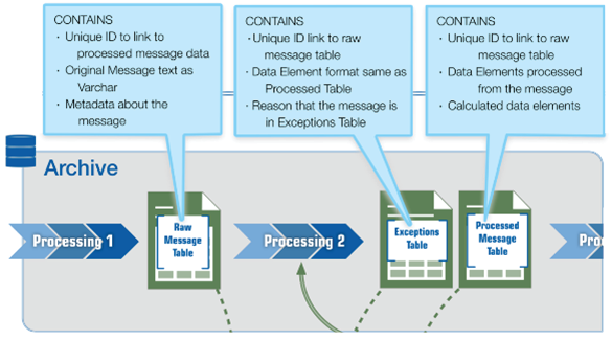

Many analysts and epidemiologists are accustomed to performing validation and quality improvement activities on their site’s incoming syndromic data—most often on the Processed data but also on the Raw data. When developing the BioSense Platform, the NSSP team made sure sites would still have such capabilities. So, to accommodate quality improvement activities, the Analytic DataMart was conceived. The Analytic DataMart, or “DataMart,” resides on a server separate from all data processing into ESSENCE. Separation is maintained so that analytics and validation activities do not negatively affect data flow. This separation is made possible by a process called replication.
Replication is the process by which all data in the Archive processing lane are copied to DataMart incrementally, about every 10 to 15 minutes. All NSSP Data Quality Reports and Data Quality Dashboards are based on DataMart data. Authorized users may access these data on the BioSense_Platform using the RStudio Workbench or SAS Studio web applications. These apps may be accessed via the Access & Management Center (AMC) home page.
All tables accessible in the BioSense_Platform are replicated views that include:
- XX_MFT—site Master Facility Table
- XX_Operational_Crosswalk
- Production Views:
- XX_PR_Raw—original messages with key metadata
- XX_PR_Processed—processed messages with calculated values that meet minimum processing criteria
- XX_PR_Except—processed messages that did NOT meet minimum criteria
- XX_PR_Except_Reason—Message_IDs with all reasons for being placed in exceptions
- XX_Cache_ER_Base—one record per visit, as available in ESSENCE
- Production Onboarding (Staging) Views:
- XX_ST_Raw—original messages with key metadata
- XX_ST_Processed—processed messages with calculated values that meet minimum processing criteria
- XX_ST_Except—processed messages that did NOT meet minimum criteria
- XX_ST_Except_Reason—Message_IDs with all reasons for being placed in exceptions
- Filter_Reasons—Filter Reason Code definitions
- Except_Reasons—Exception Reason Code definitions
Note: XX refers to the site’s short name.
These tables are located on a SQL server. Access is standard ODBC (open database connectivity) via RStudio Workbench or SAS Studio with the initial data pull in SQL. Because of this, you can run analytics using a combination of SQL and R, or SQL and SAS. Basic R and SAS queries for running SQL queries on the BioSense_Platform have been provided in previous NSSP Updates (December 2020, March 2022). Please contact the NSSP Service Desk for assistance with quality improvement code. Or visit the NSSP Technical Resource Center for links to user manuals and quick start guides.
Identifying When Replication is Broken
If you’re using RStudio Workbench or SAS Studio to monitor data and don’t see recent data, look at these data in ESSENCE. If you see recent visits in ESSENCE that aren’t reflected in the data flow, it is likely that there are issues with replication. Keep in mind that replication is often up to 15 minutes behind—and, at times, ESSENCE can be up to 4 hours behind. If you see evidence of problems, please submit an NSSP Service Desk ticket.
Getting Access to RStudio Workbench and SAS Studio*
Contact your site administrator and request access. Site administrators, please make sure the requestor has an account set up in the AMC. Once the user account is set up, grant access in the following ways:
- DataMart: Edit the User Profile within the AMC. Check the box next to DataMart.
- SAS Studio: Edit the User Profile within the AMC. Check the box next to SAS Studio.
- RStudio Workbench: Submit an NSSP Service Desk ticket requesting that the user be given access to RStudio.
* Note: Granting access to DataMart and using either analytic tool will allow access to line-level data received from all facilities within your site

The Data Flow Journey
If you found this article helpful, check out the others in NSSP’s Technical Resource Center, Onboarding Articles.


◾ 4,400,000 from Nonfederal EDs and Urgent Care Facilities
◾ 400,000 from U.S. Department of Defense
◾ 100,000 from U.S. Department of Veteran Affairs
◾ 1,100,000 from Commercial Laboratory (2 Labs)
