Ten Years of Gains: A Look Back at Progress Since the 2009 H1N1 Pandemic
June 11, 2019 — In 2009, a novel H1N1 influenza (flu) virus emerged to cause the first flu pandemic in 40 years. The 2009 H1N1 pandemic was estimated to be associated with 151,700 to 575,400 deaths worldwide during the first year it circulated. [1] This H1N1 virus has continued to circulate seasonally to this day. CDC and its many partners have made great strides in the fields of influenza surveillance, prevention, and treatment since 2009, benefiting both the annual response to seasonal flu epidemics, as well as the global capacity to respond to the next pandemic. Key improvements are summarized on the pages below.
When the pandemic H1N1 virus emerged in April 2009, laboratories were quickly overwhelmed by sharp increases in testing demands.
CDC moved quickly to expand domestic and global capacity to detect the virus by using its genetic sequence data to update existing rRT-PCR test kits.
Less than two weeks after the novel H1N1 virus was identified, revised CDC rRT-PCR test kits began shipping to public health laboratories through an FDA Emergency Use Authorization (EUA). Once labs had the CDC test kit and verified that their testing was running properly, they were able to process their own specimens and no longer needed lab confirmation from CDC.
Gene sequencing technology
During the pandemic, owing in part to the preparedness work done prior to 2009, CDC and public health laboratories were able to use molecular testing technology, “with its pinpoint accuracy and revolutionary speed” [2] to detect cases, and monitor the spread of the virus and its characteristics, including watching for emerging drug resistance, for example. In the wake of the 2009 pandemic, numerous retrospective analyses deemed the use of this technology to be one of the response’s key successes [3].
Since the pandemic, CDC’s ability to map the complete influenza virus genome has improved exponentially, thanks to considerable leaps forward in gene sequencing technology, sometimes referred to as Next Generation Sequencing or Advanced Molecular Detection (AMD) technology. Where previous technologies revealed the genes of the predominant influenza virus in a respiratory specimen, it’s now possible to see the gene sequences of all of the influenza particles in a single specimen, thereby giving deeper insight into how influenza viruses may change, for example, by mutating inside a single patient to become resistant to antiviral drugs.
In 2012 CDC’s influenza laboratory transitioned from a characterization-first approach to a sequence-first approach so now all flu viruses undergo full sequencing as a first step when they arrive at CDC. This change has reduced public health response time to flu outbreaks and also served to greatly expand the global repositories of influenza gene sequence data. Meanwhile, ongoing technological improvements by device manufacturers and CDC’s innovative protocols have brought the gene sequencing cost per virus down from about $180.00 in 2012 to $25.00 per virus in 2019, with additional improvements in the works to bring the cost down even further.
In 2009, CDC sequenced influenza virus genomes primarily for the identification of vaccine reference viruses. Now, CDC performs “next generation sequencing” on close to 7,000 influenza viruses annually, and has submitted more than 30,000 flu virus genomes to public databases.
Influenza public health laboratory tests
RT-PCR
Since 2009, the widespread adoption by public health laboratories of the testing technique called “real-time Reverse Transcription-Polymerase Chain Reaction” (rRT-PCR) with CDC’s flu rRT-PCR test kits has directly enhanced the nation’s pandemic preparedness in a number of ways. For one, widespread use of both has led to standardization of influenza testing across the nation’s public health laboratories. Relative to the other diagnostic methods widely used by laboratories to reveal the type and subtype of flu virus in a sample, rRT-PCR produces more-reliable results, and produces those results faster than most other laboratory techniques. [4]
CDC’s primary rRT-PCR test for influenza viruses (called the ‘CDC Human Influenza Virus Real-Time RT-PCR Diagnostic Panel’) is an internationally recognized reference method for detection of influenza. This means the performance of other detection methods is often measured against the performance of CDC’s rRT-PCR flu test.
The adaptability of the rRT-PCR test allows laboratories to quickly adjust how specimens are processed in outbreak and pandemic situations to avoid backlogs and unnecessary use of resources. CDC provides algorithms that help to ensure that as more specimens are tested, reagents are conserved, thereby maximizing their public health benefit and further reducing the possibility of supply shortages.
Although laboratories have the option of using other rRT-PCR tests, use of the CDC test in lieu of commercially manufactured rRT-PCR tests takes the pressure off individual laboratories of ensuring their tests are able to detect the newest emerging viruses.
CDC uses gene sequence data to update its influenza diagnostic kits and reagents, which are used around the world by public health laboratories as the gold-standard for detecting influenza, in large part because of CDC’s rapid response in updating the kits and reagents each time a novel virus emerges.
For example, in 2012 the U.S. experienced a rapid up-tick in human infections with swine influenza viruses (called variant virus infections) associated with exposure to infected pigs. CDC quickly confirmed the CDC rRT-PCR test kit’s ability to detect those swine viruses appearing in people, and then issued guidance to laboratories on how to interpret rRT-PCR test results when testing specimens from patients with known pig exposure. Since then, CDC has monitored how the diagnostic kit has performed in detecting variant infections and has updated guidance and virus-specific assay materials (called reagents) to make sure the tests are able to detect these viruses as they have evolved.
In April 2013, shortly after the first influenza A(H7N9) human infections in China were reported and within days of China CDC sharing gene sequences of the H7N9 virus, U.S. CDC quickly modified and then quality-checked CDC’s existing H7 rRT-PCR test and drafted protocols and guidance for its use. On April 22, 2013, the U.S. Food and Drug Administration (FDA) issued an Emergency Use Authorization and the H7 rRT-PCR test with test components and CDC guidance were made available to public health laboratories so they would be able to test for H7N9 viruses too.
International Reagent Resource
While RT-PCR is fast and accurate, it requires a steady stream of laboratory supplies called reagents (primers, probes and enzymes) to perform testing, which can become scarce or costly during outbreaks when testing demands increase.
To address this potential shortcoming, in 2008 CDC established the Influenza Reagent Resource (IRR) as a warehouse for CDC-developed influenza supplies that laboratories need to carry out basic research; develop diagnostics, vaccines and drugs; and conduct surveillance for emerging influenza threats. During the pandemic, the IRR proved to be a critical resource in terms of providing domestic and international laboratories with the supplies needed to test for the new 2009 H1N1 virus.
Ten years later, the IRR is firmly established as a central component of influenza research, surveillance and diagnostic testing in the United States. Further, it also has been expanded to include production and distribution of additional CDC-developed reagents for a wider range of pathogens that are within the scope of the National Center for Immunization and Respiratory Diseases (NCIRD).Some of the newer additions include reagents for respiratory syncytial virus (RSV), Middle Eastern Respiratory Syndrome (MERS-CoV), meningitis, and Streptococcus pneumoniae. The program’s broader scope is reflected in a new name, the International Reagent Resource.
With regard to pandemic preparedness, public health laboratories across the globe continue to use the reagents they receive from the IRR for surveillance of novel influenza viruses (such as H7N9 and H5N1) in tandem with the World Health Organization (WHO) Global Influenza Surveillance and Response System (GISRS). For influenza and other pathogens, CDC’s subject matter experts provide technical guidance and oversight to ensure that IRR acquires, authenticates, manufactures and distributes quality-controlled reagents that support global surveillance efforts.
Advances in Tests Used in Clinical Settings

Compared to ten years ago, clinicians now have more tests available for detection of influenza viruses in respiratory specimens, including a wider selection of highly accurate molecular assays (some rapid, some not), and improved rapid influenza antigen detection tests (RIDTs).
Rapid and accurate diagnosis of influenza virus infection facilitates timely patient management for seasonal influenza as well as pandemic influenza. Influenza testing has been used to inform decisions on the use of antiviral drugs for treatment, to avoid misuse of antibiotics for treatment, and to reduce the need for other diagnostic tests. Influenza testing also can be helpful in informing recommendations for sick people living with others who are at high risk of developing serious influenza complications. See Guide for considering influenza testing when influenza viruses are circulating in the community for more information.)
The need for better rapid influenza tests and clear rapid test guidance for clinicians became apparent during the 2009 H1N1 pandemic, when false-negative rapid antigen detection test results contributed to delays and missed opportunities in treating pandemic flu virus-infected patients with antiviral drugs; and delays in implementing infection control measures for such patients. [5]
Shortly after the pandemic, CDC and partners began working to address issues with rapid antigen detection tests available at the time, reaching an important milestone in 2017 when rapid influenza antigen detection tests (RIDTs) were reclassified by FDA (described in more detail, below) and held to higher standards.
In June 2014, a new kind of rapid influenza test was approved by FDA (the Alere (TM) i Influenza A&B test by Alere Scarborough, Inc D/B/A Binax, Inc, now owned by Abbott and called the ID NOWTIM Influenza A&B) which detects flu A and B viruses by detecting the PB2 flu virus gene in respiratory specimens. This approval marked the beginning of a new category of tests, referred to as rapid influenza molecular assays.
Rapid influenza molecular assays are a relatively new type of influenza diagnostic test. These tests are similar to RT-PCR (which is also a molecular process and currently the gold standard for influenza virus detection), in that both tests use nucleic acid amplification, which detects influenza viruses in a respiratory specimen by amplifying (multiplying) certain nucleic acids (building blocks of genes) in the influenza virus genome.
Since 2014, other rapid influenza molecular assays have been approved by FDA and are available for use in clinical settings. Of those, some are approved for point-of-care or bedside use, and do not require a clinical laboratory, including Cobas® Influenza A/B and Cobas® Influenza A/B RSV Assay by Roche Molecular Diagnostics; Xpert Xpress Flu and Xpert Xpress Flu/RSV by Cepheid; Accula Flu A/Flu B assay by Mesa Biotech Inc,; and ID Now TM by Abbot.
Rapid influenza molecular assays have a strong ability to identify correctly patients with evidence of influenza virus infection (referred to as high sensitivity, greater than 90%) and a strong ability to identify correctly patients without evidence of influenza virus infection, referred to as high specificity (90-100%). These assays produce results in 15 to 30 minutes, making them a convenient option for clinical management of patients. Other rapid influenza molecular assays (such as the Cobas® Influenza A/B & RSV Assay by Roche Molecular Diagnostics) are approved that detect influenza A and B viruses and respiratory syncytial virus (RSV) in respiratory specimens. These tests are particularly useful in the clinical management of young children with acute respiratory illness, but can be used for persons of all ages.
Additionally, a number of other influenza molecular assays are FDA-approved, for use in a moderately complex or complex hospital clinical laboratory. These assays may take more than one hour to produce results. Many of these molecular assays detect influenza A and B viruses as well as other respiratory viruses such as adenovius, coronavirus, human metapneumovirus, human rhinovirus/enterovirus, parainfluenza virus, and RSV. Some also detect some respiratory bacterial infections.
Both rapid molecular and other molecular assays are more accurate than previously available influenza tests for use in clinical settings. These advances in molecular technology provide more-accurate influenza testing results and are likely to improve clinical management of patients with suspected influenza in ambulatory care clinics and emergency departments, and in hospitalized patients as well.
With regard to rapid influenza antigen detection tests (RIDTs), to address the issues identified during the pandemic, in 2011 the CDC, the Joint Commission, the Biomedical Advanced Research and Development Authority (BARDA), the Medical College of Wisconsin (MCW) and other public and private partners addressed key RIDT-related issues by:
- Creating the first method for systematically evaluating commercially available RIDTs, described in a 2012 Morbidity and Mortality Weekly Report (MMWR) article
- Enhancing awareness among clinicians of appropriate RIDT protocols with new courses, videos and decision-making tools, and;
- Working with FDA to reclassify RIDTs from Class I to Class II with Special Controls, thereby holding current and new RIDTs to higher performance standards.
Reclassification means that RIDTs are now subject to the following requirements [6]:
- Manufacturers must test their RIDTs annually to ensure they can detect currently circulating seasonal flu viruses. RIDTs with lower sensitivity to those flu viruses will need to indicate so on their labeling.
- RIDTs must meet minimum performance criteria, such as high sensitivity and high specificity to detect flu viruses in respiratory specimens compared to RT-PCR or viral culture. This means that currently FDA-approved RIDTs will now be more accurate in detecting flu viruses in respiratory specimens than previous RIDTs.
- In the event of a pandemic, manufacturers must test the reactivity of their RIDTs with the newly emergent flu virus as soon as virus samples become available.
- Since 2017 when reclassification of RIDTs occurred, manufacturers have made positive, steady changes to rapid influenza antigen detection tests, but more work still needs to be done.
Surveillance
Right-sizing initiative

The massive amount of laboratory testing that occurred during the 2009 H1N1 pandemic provided an opportunity for researchers to identify the optimal levels of influenza surveillance and laboratory testing needed in the United States, including ways to improve efficiency.
To answer those questions, in 2010, CDC and the Association of Public Health laboratories (APHL) began developing a “right-size” approach to influenza virologic surveillance which was based on extensive input from public health laboratories and stakeholders. Since then, this right-size approach has helped public health laboratories to:
- standardize virologic surveillance practices, determine the optimal number of specimens to test to produce statistical confidence in resulting data, and define public health surveillance priorities;
- adopt requirements, resources and statistical calculators that aid in planning and justifying budget and resource requests;
- increase the understanding and support from political leaders and the public;
- speak a common language between laboratories and epidemiologists; and
- assist decision makers in analyzing the impacts of budget decisions on national surveillance objectives, especially with regard to pandemic preparedness capacity.
Automated transmission of laboratory reports
In addition to identifying the optimum “sampling” strategy, major improvements in sharing this data have taken place. In March 2010, five laboratories were routinely sharing data electronically with CDC. Today, all public health laboratories at the state level and some local public health and clinical laboratories send data electronically to CDC’s Influenza Division This increase has improved the timeliness and completeness of reporting, for both seasonal influenza surveillance activities and the identification of novel influenza A viruses.
Incorporation of NCHS mortality data
In collaboration with the National Center for Health Statistics (NCHS), since the 2015-2016 flu season CDC’s Influenza Division has used mortality data collected from death certificates and reported to NCHS as the principal tool to track influenza-associated mortality in the United States.
The shift to NCHS data marked a significant advance in the capacity to track flu mortality, owing in part to efforts made by NCHS to improve the timeliness of jurisdiction reporting and modernize the infrastructure of national vital statistics, all of which resulted in a system capable of supporting near real-time surveillance of most deaths in the United States.
The new system presents deaths by the date of occurrence rather than by the date on which deaths were registered. It also provides a consistent case definition and covers nearly all deaths occurring in the United States, which is an improvement over the previous (now retired) system which accounted for only 25 percent of all US deaths.
Inclusion of electronic data in ILINet
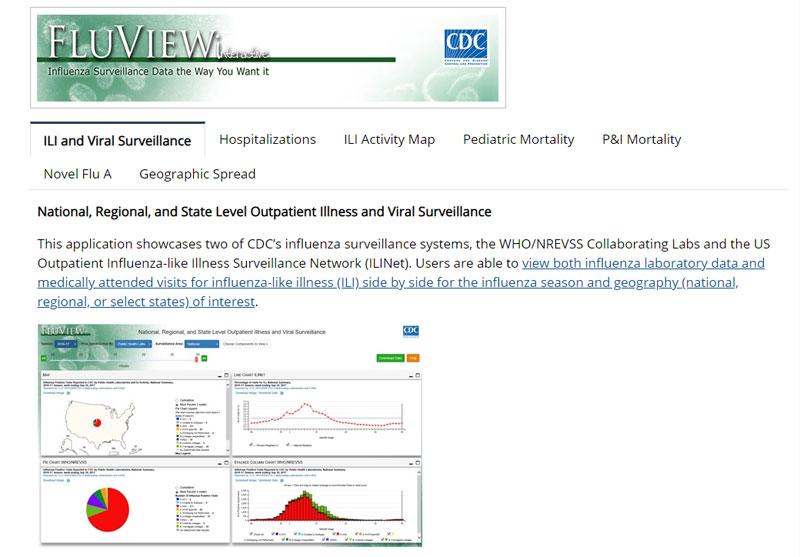
CDC’s U.S. Outpatient Influenza-Like Illness Surveillance Network (ILINet) collects data each week from up to 3,500 outpatient health care providers across all 50 states, Puerto Rico, the District of Columbia and the U.S. Virgin Islands. Each week, these health care providers report the total number of patients seen, and the number of patients seen with influenza-like illness (ILI), by five age groups. The process of reporting numbers of total patients and ILI patients was labor-intensive, sometimes leading to reporting delays and dips in participation. Since the pandemic, more ILINet providers have used electronic health records in determining ILI and patient visits, thereby reducing the reporting burden and providing more-comprehensive, efficient and timely surveillance. ILINet now captures more patient visits: during the 2018-19 season, more than one million patient visits were reported weekly, almost double the number of patient visits reported weekly during the 2009 pandemic.
Antiviral resistance testing
Another step forward in surveillance is the number of public health laboratories in the U.S. that test for resistance to influenza antiviral medications, which increased from four laboratories before the 2009 H1N1 pandemic to 20 laboratories in 2019. This improvement was accomplished through training courses, site visits, technical and other support from CDC. In addition, four laboratories are now able to conduct advanced testing for antiviral drug resistance, using a more sensitive and complex procedure. Since 2018, CDC’s influenza laboratories have been testing seasonal and animal viruses for susceptibility to the newest anti-influenza drug – baloxavir marboxil (Xofluza) and a CDC-developed assay is now being implemented at the public health laboratory designated for antiviral testing (NIRC, Albany NY). Lastly, in collaboration with a World Health Organization (WHO) expert group, reference laboratory materials for detection and reporting of influenza drug resistance have been developed and shared with laboratories around the world.
The Burden of Influenza
During the pandemic, influenza hospitalization rates increased, particularly among groups of people at high risk of developing serious flu complications.
The Influenza Hospitalization Surveillance Network (FluSurv-NET) conducts population-based surveillance for laboratory-confirmed influenza-related hospitalizations in children and adults. It has provided critical data for:
- informing policy and decision making, especially with regard to vaccine and antiviral prioritization;
- evaluating the effectiveness and safety of pandemic vaccine; and,
- identifying groups at high risk for severe flu complications.
Over the course of the pandemic, CDC refined and revised its hospitalization surveillance methods, eventually developing a way to use FluSurv-NET data to estimate the range of 2009 pandemic flu illnesses, hospitalizations and deaths (also referred to as ‘estimates of flu burden’) in the United States.
Since the 2009 pandemic, this method of estimation has been applied at the end of each flu season as a means of illustrating the national impact of seasonal flu. In 2019, CDC also began providing on a weekly basis preliminary estimates of the burden of influenza during flu season.
Burden of influenza averted through vaccination

CDC also developed estimates of the number of flu illnesses, hospitalizations and deaths prevented by flu vaccination each flu season. These estimates are made with mathematical models that combine illness rates, vaccine coverage, and vaccine effectiveness, to estimate the amount of flu that would have occurred without flu vaccination. The amount of flu that was prevented by vaccination was the difference between what would have occurred and what was estimated to have actually occurred. CDC has continued to estimate the benefits of influenza vaccination each season. Now, CDC and its partners are better able to communicate the benefits of flu vaccination and provide clear evidence on the public health impact that vaccination can have in reducing flu illness.
Vaccine effectiveness monitoring
While FluSurv-NET provided vaccine effectiveness (VE) data during the 2009 H1N1 pandemic, the US Flu VE Network has yielded in-depth data about how well flu vaccines are working each flu season since its inception in 2005.
Seasonal VE data are instrumental in making vaccine policy decisions and improving virus selection to update the composition of seasonal vaccines. VE data are also pivotal to understanding the epidemiology and severity of pandemic and seasonal influenza and to mounting an appropriate responses.
Following the pandemic, multiple improvements were made to the US Flu VE Network including expanding the number of sites, increasing enrollment, improving the timeliness of data collection and reporting, and building an infrastructure for conducting special studies to improve the benefits of vaccines. In addition, in 2016-17 CDC established the Hospitalized Adult Influenza Vaccine Effectiveness Network (HAIVEN), which focuses on VE among severely ill adults, as opposed to VE among persons seen as outpatients.
With regard to available data, at the time of the pandemic, VE studies had shown that influenza vaccination reduced the risk of mild or moderately severe influenza by approximately half among children and adults, but there were gaps in knowledge about how well the vaccine worked in preventing lab-confirmed influenza among pregnant women and in preventing life-threatening influenza among children and adults.
VE data directly influence policymakers’ decisions regarding influenza vaccination policy both in the United States and in other countries, so over the last 10 years CDC has created partnerships and funded research to address major VE knowledge gaps, all with an eye toward advancing vaccination efforts in the U.S. and internationally.
Pregnant Women
The first CDC-funded study to address these gaps, which was also the first to use laboratory-confirmed influenza outcomes to assess VE among pregnant women, was published in the journal Clinical Infectious Diseases in January 2014. This study, which looked at Kaiser Permanente health plan members in two regions, showed that influenza vaccination reduced their risk of flu illness by half during 2010 to 2012, indicating that flu vaccine works about as well in pregnant women as in other young, healthy adults. The study showed influenza vaccine effectiveness estimates among pregnant women were similar to those found in other studies among young adults, which ranged from 44% to 51% from 2010 to 2012.
The second CDC-funded study was published in Clinical Infectious Diseases in October 2018: it was the first study to show that vaccination protects pregnant women against flu-associated hospitalization.
To conduct this study, CDC partnered with a number of other public health agencies and health care systems in Australia, Canada, Israel, and the United States through the Pregnancy Influenza Vaccine Effectiveness Network (PREVENT), which consists of health care systems with integrated laboratory, medical, and vaccination records. Sites retrospectively examined medical records of more than two million women who were pregnant from 2010 through 2016 to identify those who were hospitalized with laboratory-confirmed flu.
Key findings of this study include the following:
- Over the course of six flu seasons, getting a flu shot reduced a pregnant woman’s risk of being hospitalized from flu by an average of 40 percent.
- More than 80 percent of pregnancies overlapped with flu season, underscoring the likelihood that pregnant women will be exposed to flu at some point during their pregnancy.
- Flu vaccine was equally protective for pregnant women with underlying medical problems such as asthma and diabetes, which also increase the risk of serious medical complications including a worsening of those chronic conditions.
- Flu vaccine was equally protective for women during all three trimesters.
Children and Adults:
In March 2014 another critical gap was filled, when the first study to estimate VE in children against flu admissions to pediatric intensive care units (PICUs) was published in the Journal of Infectious Diseases. CDC researchers found that getting a flu vaccine reduces a child’s risk of flu-related intensive care hospitalization by 74%, illustrating the important protection flu vaccine can provide to children against more-serious flu outcomes.
Researchers analyzed the medical records of 216 children age 6 months through 17 years admitted to 21 PICUs in the United States during the 2010-2011 and 2011-2012 flu seasons.
Though flu vaccination was associated with a significant reduction in risk of PICU admission, flu vaccine coverage was relatively low among the children in this study: only 18 percent of flu cases admitted to the ICU had been fully vaccinated.
More than half (55 percent) of PICU cases had at least one underlying chronic medical condition that placed them at higher risk of serious flu-related complications.
See New Study Shows Flu Vaccine Reduced Children’s Risk of Intensive Care Unit Flu Admission by Three-Fourths for more information.
In August 2018, a CDC-supported study published in Vaccine was the first to provide statistically significant VE estimates for adults against hospitalization and admission to ICU.
The study was conducted over four flu seasons from 2012 to 2015 and found that flu vaccination prevented severe disease:
- Flu vaccination among adults reduced the risk of being admitted to the hospital with flu and placed in a general ward bed by 37 percent.
- Flu vaccination was even more effective in preventing the most severe forms of flu and reduced the risk of being admitted to an ICU with flu by 82 percent.
Because flu vaccine varies in how well it works and people who are vaccinated may still get sick, the study also looked at whether flu vaccination reduced the severity of illness among hospitalized people who were vaccinated compared to those who were unvaccinated and found that:
- Among adults who were admitted to the hospital with flu, vaccinated adults were 59 percent less likely to have very severe illness resulting in ICU admission than those who had not been vaccinated.
- Among adults in the ICU with flu, vaccinated patients on average spent 4 fewer days in the hospital than those who were not vaccinated previously.
The study was a collaborative project with CDC, conducted through the Southern Hemisphere Influenza and Vaccine Effectiveness Research and Surveillance project, which prospectively enrolled hospitalized adults 18 years and older from 2012 to 2015 in Auckland, New Zealand. Eligible hospitalized patients were those who had an overnight admission with acute respiratory illness. Once enrolled in the study, patients self-reported their flu vaccination status and were tested for flu infection by RT-PCR.
See Study Shows Flu Vaccine Reduces Risk of Severe Illness for more information.
In February 2018, the complex process of making CVVs for avian H5 and H7 influenza viruses was streamlined by the Federal Select Agent Program with its decision to remove the requirement for live bird lethality testing.
H5 and H7 avian influenza viruses are classified as Select Agents until it is shown that the viruses are low-pathogenic (unable to cause disease and mortality in chickens in a laboratory setting).
Previously, live bird lethality testing was used to determine whether an avian influenza CVV was highly pathogenic or low pathogenic, and thus if it should managed as a Select Agent.
Now, H5 and H7-derived CVVs no longer need to be tested in live birds to show low pathogenicity. Instead, other data may be submitted in lieu of bird lethality testing.
The Federal Select Agent Program is managed jointly by the Division of Select Agents and Toxins at the CDC, and the Agriculture Select Agent Services at the Animal and Plant Health Inspection Service.
Prevention
Influenza Vaccines
CDC and its partners have made significant progress in influenza surveillance; diagnostics; characterizing viruses for vaccine strain selection; and developing systems to evaluate the effectiveness of influenza vaccines over the last ten years.
For example, with regard to vaccine development, by using newer production technologies CDC can now identify and provide candidate vaccine viruses for novel influenza threats to manufacturers within a matter of weeks.
This and other improvements have helped to better protect the public from seasonal and pandemic influenza threats through vaccination. However, to more fully protect Americans from seasonal and pandemic flu, more effective vaccines are needed and more people still need to receive annual flu vaccines.
In line with that public health mission, more doses of seasonal vaccines and different vaccine products are available than ever before. In addition to trivalent inactivated vaccine and live attenuated influenza vaccine, the following vaccines have been approved by the FDA and are now available:
- a high dose vaccine that is designed specifically for people 65 and older to help create a stronger antibody response;
- a trivalent flu vaccine made with adjuvant (an ingredient added to vaccine that helps create a stronger immune response), which was approved for people 65 years of age and older;
- the first U.S.-approved cell-based flu vaccine, which can potentially be made more quickly than traditional egg-based vaccines and does not require a large supply of eggs to produce;
- quadrivalent (four component) flu vaccines that protect against both lineages of influenza B viruses thus offering expanded protection against circulating influenza viruses; and,
- recombinant influenza vaccines, which do not require an egg-grown vaccine virus or eggs to produce, and which may be manufactured more quickly than egg-based vaccines.
CDC is currently exploring new ways to further improve influenza vaccine through the influenza vaccine improvement initiative (iVii). The initiative includes two primary goals.
Goal 1: Build the evidence base for developing more-effective influenza vaccines, and increase the impact of vaccines that are currently available.
This goal points to the need for deeper data, so during the 2018-19 season, CDC increased the number and scope of VE Network participants by over 1,500 children and adults, bringing the total number of participants enrolled to more than 10,500.
CDC also is increasing the diversity of people who can be enrolled in studies and has expanded VE monitoring through innovative use of health care and other data sources outside of the US Flu VE Network. The laboratory process of evaluating vaccine response through the use of enhanced serologic and cellular testing has also been improved.
Goal 2: Increase the capacity of CDC laboratories to select, develop, evaluate and perform virus characterization to provide candidate vaccine viruses.
To accomplish this, CDC is employing state-of-the-art technologies to increase the volume of laboratory testing being done. CDC also is working on developing new assays for manufacturers and regulatory laboratories, and planning evaluation projects that support vaccine improvement.
CDC also is focusing on expanding and improving global virus detection, and improving vaccine effectiveness monitoring. This is being done through the expansion of Next Generation Sequencing and fully transitioning to the Sequence-First initiative described earlier. In the process, CDC has also worked with partners to automate the pipeline used to produce, store and share the enormous volume of NGS data.
CDC also is piloting an advanced laboratory strategy to identify viruses, using antigenic data that are likely to predominate in the human population in future influenza seasons.
CDC meets regularly with vaccine manufacturers and other WHO Collaborating Centers and Essential Regulatory Laboratories including FDA’s Center for Biologics Evaluation and Research to share information on a number of vaccine-related topics.
Topics include the availability of candidate vaccine viruses for use in the development and production of seasonal influenza vaccines, the availability of protocols and reagents needed for the development, standardization, and regulation of influenza vaccines, and to discuss potential issues related to the timely production of seasonal influenza vaccines. The group meets from the time the vaccine composition is announced until vaccines are released for distribution to health care providers, for both the northern and southern hemisphere influenza seasons.
Separately, frequent Flu Risk Management Meetings serve as a venue to discuss issues relating to the U.S. Department of Health and Human Services (HHS) response to seasonal and pandemic influenza. Subjects for discussion include but are not limited to seasonal surveillance updates, effectiveness of influenza vaccines, vaccine and antiviral stockpiles, emerging influenza virus surveillance, clinical trial response to influenza outbreaks of novel influenza viruses, and pandemic preparedness.
When a pandemic influenza A virus emerges – like H1N1 in 2009 – well-matched vaccines may not be available for 6 months or longer, and antiviral medications may be reserved for treatment and be in short supply because of high demand.
The goals of community mitigation measures are to delay the spread of the disease and reduce the impact of an influenza pandemic in U.S. communities.
Community mitigation measures are often referred to as Non Pharmaceutical Interventions (or NPIs), and include actions other than use of vaccines and medications that people and communities can take to help slow the spread of a pandemic influenza virus.
Community Mitigation Measures
Community Mitigation Guidelines to Prevent Pandemic Influenza – United States, 2017
Based on lessons learned from the 2009 H1N1 pandemic response and an expanded contemporary NPI evidence base, CDC published updated pre-pandemic planning guidelines – Community Mitigation Guidelines to Prevent Pandemic Influenza – United States, 2017 – in MMWR Recommendations and Reports on April 21, 2017. The guidelines offer free CE credits for healthcare and public health practitioners, and are available at https://www.cdc.gov/mmwr/volumes/66/rr/pdfs/rr6601.pdf.
The 2017 guidelines summarize key lessons learned from the 2009 H1N1 pandemic response; encourage state and local public health officials to plan and prepare for implementing NPIs early in an influenza pandemic in community settings; describe new or updated pandemic planning and assessment tools; and provide the latest scientific findings to support updated recommendations on the use of NPIs to help slow the spread and decrease the impact of an influenza pandemic. The 2009 H1N1 pandemic response highlighted that pre-pandemic planning and preparedness – from local to federal levels – must be broad, flexible, and multi-sectoral, and emphasized the critical value of public engagement and community preparedness – including families, schools, and businesses – for successful NPI implementation during a pandemic. Research conducted during and in the wake of the 2009 H1N1 pandemic documented that public acceptance of NPIs in the United States is generally high, even for disruptive measures like school closures. These findings underscore the importance of pandemic preparedness at all levels of society to ensure timely implementation of community mitigation measures from the onset of a future pandemic, when these measures may be the only tool available in many jurisdictions.
To help implement the 2017 guidelines and to assist states and localities with pre-pandemic planning and decision-making in their communities, CDC also published online six plain-language, pre-pandemic NPI planning guides for various audiences and community settings: households, educational settings, workplace settings, community- and faith-based organizations serving vulnerable populations, event planners of large/mass gatherings, and health communicators in community preparedness. The six guides are available.
CDC continues to conduct and support research to assess the feasibility, acceptability, and effectiveness of NPI implementation in various community settings. Select NPI research references, as well as additional NPI communication, education, and training materials, are available at CDC’s dedicated NPI website.
Treatment

The number of available, approved and recommended treatment options has increased in the last ten years.
During the 2009 H1N1 pandemic, the influenza antiviral medication oseltamivir (oral oseltamivir, available under the trade name Tamiflu®) was used extensively for treatment, while zanamivir (inhaled, trade name Relenza®) was used less.
Oral oseltamivir’s widespread use was due to it being approved, recommended and utilized for treatment of patients hospitalized with severe influenza. It also was recommended for use in hospitalized patients with non-severe influenza, although no antiviral medications were approved by FDA for use in that group.
Outpatients could be prescribed either oral oseltamivir or inhaled zanamivir, which were both approved for early treatment of uncomplicated influenza by FDA.
Of the two medications, CDC was more concerned with the possibility of resistance emerging against oseltamivir, which did happen, but not often. The oseltamivir-resistant viruses that did emerge were not transmitted easily from person to person, and zanamivir was used effectively to treat them.
Beginning in April 2009 and continuing for a few years after the pandemic, the FDA’s Emergency Investigational New Drug (EIND) program provided an application process which authorized investigational use of IV zanamivir for patients with severe and life-threatening influenza.
Later, on October 23rd, 2009, FDA issued an Emergency Use Authorization (EUA) for IV Peramivir. At the time, IV Peramivir was an investigational intravenous antiviral drug used rarely to treat people who had been hospitalized with severe influenza. The drug was held in the Strategic National Stockpile (SNS) and distributed by CDC under the EUA. Licensed clinicians were able to request this product through the CDC website electronic request system, and product was delivered directly to hospital facilities until June 2010 when the EUA was terminated.
Following the pandemic, in December 2014, IV Peramivir was approved by FDA for early treatment of uncomplicated influenza in outpatients, which also opened up some off-label use in treating influenza in hospitalized patients. Previously, with the exception of the IV Peramivir EUA during the pandemic, oseltamivir and zanamivir were the only recommended antiviral medications for treatment of influenza. (The other approved antiviral drugs – amantadine and rimantadine – were not and are still not recommended due to high levels of resistance detected in circulating influenza viruses).
Roughly three years later, in September 2017, FDA approved the first generic version of oseltamivir.
Oseltamivir, zanamivir and IV peramivir are all neuraminidase-inhibitor influenza antiviral medications, so named because each works by targeting the neuraminidase surface protein of the influenza virus to stop the virus from being released from infected cells and spreading to healthy cells.
In December 2018, a new influenza antiviral medication called oral baloxavir marboxil (trade name Xofluza®) was approved by FDA and is recommended for treatment of influenza. Baloxavir works differently, primarily by preventing an influenza virus from multiplying when it is inside a cell. Because baloxavir works differently, it is in a new class of antiviral medications called cap-dependent endonuclease inhibitors. Like the neuraminidase-inhibitor medications, baloxavir has activity against both influenza A and B viruses. Baloxavir is approved for early treatment of uncomplicated influenza in outpatients aged 12 years and older.
With more antiviral medications approved, recommended and available, treatment options have improved for both hospitalized patients with severe influenza and outpatients seeking treatment early for uncomplicated influenza. There remains a gap, however, in approved antiviral treatment options for hospitalized patients with non-severe influenza, although CDC and IDSA (Infectious Diseases Society of America) continue to recommend that those patients be treated with neuraminidase-inhibitor antiviral medications.
Decision-making tools
Risk assessment (IRAT)
The 2009 H1N1 pandemic highlighted the public health value of developing an objective, scientifically based tool for assessing the potential pandemic risk posed to humans by influenza A viruses circulating in animals. To fill that need, CDC developed an evaluation tool now called the Influenza Risk Assessment Tool (IRAT) with help from global animal and human health influenza experts. IRAT launched in 2011 and since then CDC has used it to evaluate the potential risk posed by viruses that are not currently circulating in people. The IRAT relies on input from subject matter experts representing a variety of expertise in the study of influenza viruses. It uses 10 evaluation criteria grouped into major categories including properties of the virus, attributes of the human population, and epidemiology and ecology of the virus, to generate scores that indicate the potential risk of the virus to emerge as a pandemic virus, and the potential impact if it does.
The IRAT is not a prediction tool. Rather, the IRAT provides structure to prioritize and maximize investments in pandemic preparedness; identify key gaps in information; document transparently the data and scientific process used to inform management decisions associated with pandemic preparedness; provide a flexible means to easily and regularly update the risk assessment of novel flu viruses as new information becomes available; communicate effectively to the general public, policymakers, public health laboratories and other stakeholders; and provide a means to weigh the 10 evaluation criteria differently depending on whether the intent is to measure the ability of a virus to “emerge” as a pandemic-capable virus, or “impact” the human population after emerging.
Since its inception eight years ago, the IRAT has been used by CDC to evaluate and inform pandemic preparedness decisions for 16 viruses, the results of which are listed at Summary of Influenza Risk Assessment Tool (IRAT) Results.
Forecasting
Unlike traditional surveillance systems that lag behind the real-time situation, flu forecasting offers the possibility to look into the future and forecast when and where flu increases will occur, how large the impact of flu will be, and when flu will peak. Forecasts can inform messaging to health care providers regarding influenza vaccination and antiviral treatment for patients, help to prepare for an influx of illnesses and hospitalizations, and be used to guide community mitigation strategies, such as school closures.
To support the development of the science of flu forecasting and its application for public health, CDC, through the Epidemic Prediction Initiative (EPI), organized the first FluSight challenge to forecast the timing, intensity, and short-term activity of the influenza season during the 2013-14 season. Each influenza season since then, Influenza Division researchers have worked with CDC’s EPI and other external researchers to improve flu forecasting. CDC provides forecasting teams data, relevant public health forecasting targets, and forecast accuracy metrics evaluated against actual flu activity while each team submits their forecasts based on a variety of methods and data sources each week. During the 2018–19 season, forecasting teams provided over 30 national-level forecasts each week.
These challenges have provided the scientific and public health community with experience in real-time forecasting, the ability to evaluate forecast accuracy, and familiarity in communicating and applying these forecasts in real-world settings. These experiences are critical to developing a network of forecasters capable of providing results that public health officials can use, both in seasons and during an influenza pandemic.
Forecasts are currently used to inform CDC’s activity summaries provided to public health officials and CDC leadership, and to inform messages to the general public regarding the timing of the influenza season and steps the public can take to protect their health.
International work
CDC has strong global ties with other WHO Collaborating Centers for Influenza, National Influenza Centers and ministries of health around the world. These collaborators provided critical data throughout the influenza pandemic on how, when and where the pandemic virus might be changing and if the monovalent pandemic vaccine would continue to be effective in preventing infection.
CDC’s Influenza Division formed an influenza international capacity-building initiative in 2004, which provided a five-year period of financial support for nine countries to improve laboratory diagnostics and sentinel surveillance for influenza-like illness and severe acute respiratory infection. In 2009, this number increased to 37 countries receiving support under 39 cooperative agreements.
Thus, the 2009 H1N1 pandemic occurred at a time when many of the 37 countries benefited from the newly established influenza surveillance and laboratory capacities. The pandemic provided the ultimate test to determine whether their laboratory diagnostics and surveillance systems were indeed strong enough to manage the massive surges in flu activity that would come their way.
Shortly after the pandemic, eight of the 37 countries transitioned to the program’s second five-year period, called the sustainability period. During this period, financial support was reduced as the programs focused on sustaining the gains made in laboratory diagnostics and surveillance. The countries focused on standardizing foundational aspects of influenza surveillance including regular influenza activity reporting and sending viruses to CDC and other WHO Collaborating Centers, all with an eye toward ensuring preparedness for the next pandemic.
Now, following 10 years of the pandemic’s race around the globe, which caused hundreds of thousands of deaths worldwide, many countries have graduated from the sustainability period to maintenance and some are now developing in-country seasonal influenza vaccination programs based on their influenza surveillance data.
The 2009 H1N1 pandemic tested U.S. laboratory and surveillance systems and highlighted many successes along the way, shining a light on one of the biggest takeaways of the 2009 H1N1 pandemic: develop seasonal influenza epidemiology and laboratory capacity that’s flexible enough to handle the next pandemic.
1. http://www.cdc.gov/flu/spotlights/pandemic-global-estimates.htm
2. Lessons from a Virus, Association of Public Health Laboratories. https://www.aphl.org/AboutAPHL/publications/Documents/ID_2011Sept_Lessons-from-a-Virus-PHLs-Respond-to-H1N1-Pandemic.pdf
3. An HHS Retrospective on the 2009 H1N1 Influenza Pandemic to Advance All Hazards Preparedness. http://www.phe.gov/Preparedness/mcm/h1n1-retrospective/Documents/h1n1-retrospective.pdf
4. Manual for the laboratory diagnosis and virological surveillance of influenza. 2011. http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf
5. D.B. Jernigan, S.L. Lindstrom, J.R. Johnson et al. Detecting 2009 Pandemic Influenza (H1N1) Virus Infection: Availability of Diagnostic Testing Led to Rapid Pandemic Response. Clin Infect Dis. 2011 Jan 1;52 Suppl 1:S36-43. http://cid.oxfordjournals.org/content/52/suppl_1/S36.full.pdf+html
6. Refer to FDA web site for a complete list of requirements.
