AMD Activities: 2020
Updated on Tuesday, December 15, 2020

The APHL-CDC Bioinformatics Fellowship aims to train and prepare bioinformaticians to apply their expertise in public health settings where they can design tools to help public health personnel use bioinformatic data. This fellowship gives fellows at the post-masters and post-doctoral levels the opportunity to apply their skills to emerging public health issues.
Since the program’s inception in 2013, a total of 38 bioinformatics fellows have been placed in host labs at CDC or in state public health laboratories. Starting in 2016, some fellows were also placed in state public health laboratories where they started having an immediate impact. For example, the first fellow at a state public health laboratory helped increase their sequencing capacity from five different types of pathogens to over 20 different types in one year.
Each fellow brings a unique educational background and experience. In several cases, the fellowship introduced the fellow to careers in public health. To date, almost 75% of the fellows still work in public health. They also make an impact beyond their labs, serving as AMD bioinformatics resources for their regions. Collectively the fellows provide technical assistance, analysis, and training to public health laboratory scientists around the country.
Bioinformaticians can apply to be a part of the fellowship class of Fall 2021 by visiting the APHL-CDC Bioinformatics Fellowship website by February 28, 2021.
Updated on Tuesday, November 17, 2020

The Centers for Disease Control and Prevention (CDC) awarded nearly $9 million through a broad agency announcement (BAA) in fiscal year (FY) 2020 to support the SARS-CoV-2 Sequencing for Public Health Emergency Response, Epidemiology and Surveillance (SPHERES) initiative. These projects will help fill gaps in knowledge and grow the agency’s innovative approaches to respond to the COVID-19 pandemic. SPHERES builds upon the 6 years of investments from the CDC Advanced Molecular Detection (AMD) program which integrates the latest next-generation genomic sequencing technologies with bioinformatics and epidemiology expertise across CDC and the nation.
More awards to support innovative research into the COVID-19 pandemic response may be awarded in FY 2021. Funding awards are determined through a competitive selection process based on scientific merit. Through these investments, CDC will expand genomic sequencing efforts and advance public health research into the transmission and evolution of SARS-CoV-2.
Updated on Thursday, November 5, 2020

On October 27th, the NGS Quality Initiative released the next set of ready-to-implement tools and documents.
Adding to the collection of documents for Illumina Miseq, Oxford Nanopore’s MinION, and ThermoFisher’s Ion Torrent, they now offer a Bioinformatics Personnel Training SOP. There are also new training forms for CLC Genomics Workbench Employee, De novo Genome Assembly, Operating in Linux Environments, Preprocessing Read Quality Control, Read Trimming of NGS Data, and Short Read Alignment.
The July update included SOPs for training personnel on Illumina’s MiniSeq and NextSeq. Additionally, it also provided tools and documents for two new Quality System Essentials (QSEs): Information Management and Assessment. Within the release were documents on best practices for documenting bioinformatic pipelines to support reproducibility under the Information Management QSE. It also contained documents on identifying, developing, and applying key performance indicators to your sequencing assays and a Quality Management System Internal Assessment Tool to help laboratories assess compliance with quality system recommendations under the Assessment QSE.
Posted on Monday, Oct 19, 2020

Photo by: Africa Centres for Disease Control and Prevention
On October 12, a group of public and private partners led by the Africa Centers for Disease Control announced the launch of the Africa Pathogen Genomics Initiative (Africa PGI): a $100 million, 4-year collaboration to expand access to next-generation genomic sequencing tools and strengthen public health surveillance networks across the continent.
In 2015, the African Union Commission (AUC) developed the Africa Health Strategy 2016-2030 to strengthen health systems and to combat diseases in Africa. As part of this strategy, Africa PGI will build a disease surveillance and laboratory network based on genomic sequencing across 20 countries. This network will not only help identify and inform research and public health responses to COVID-19 and other epidemic threats, but also for endemic diseases such as HIV, tuberculosis, malaria, and cholera. Africa PGI will also help build lab capacity to fully utilize genomic sequencing, create a training institute for pathogen genomics, and establish an African node of the International Nucleotide Sequence Database Collaboration (INSDC) to share raw data with the international scientific community.
CDC is a contributor to the initiative and CDC’s AMD program helped to establish the Public Health Alliance for Genomic Epidemiology (PHA4GE), one of the partners that will help support the implementation of this initiative. Additional partners joining the initiative include the Bill & Melinda Gates Foundation, Illumina, World Health Organization (WHO), Oxford Nanopore Technologies, Microsoft Corporation, African Society for Laboratory Medicine (ASLM), African Academy of Science (AAS), National Public Health Institutes (NPHIs), African Centre of Excellence for Genomics of Infectious Diseases (ACEGID), and Human Heredity and Health (H3Africa).
Posted on Thursday, Sept 3, 2020
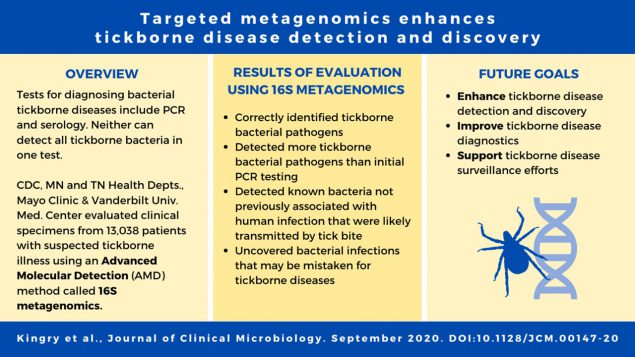
Using advanced molecular detection (AMD) techniques, CDC researchers analyzed over 13,000 leftover samples from patients who were suspected of having tickborne illness. Twelve tickborne species of bacteria that cause illness in people were detected, including two not previously associated with human illness for the presence of bacteria. A 100% increase in the number of tickborne bacteria identified by this method was demonstrated as compared to initial targeted polymerase chain reaction (PCR) assays.
Posted on Friday, August 7, 2020

One Health is an approach that recognizes the interconnectedness of the health of people, animals, and the environment. It involves collaboration across human and animal health, environment, and other relevant sectors. Antibiotic-resistant bacteria have developed the ability to defeat the drugs designed to kill them and are an important One Health issue. Today, advanced molecular detection technologies, such as whole genome sequencing (WGS), allow scientists to study how antibiotic resistance evolves and spreads. WGS also provides important information for responding to disease outbreaks. In a 2015–2018 outbreak of multidrug-resistant Salmonella Heidelberg, WGS revealed that the outbreak in people was connected to contact with dairy calves and calf environments.
Posted on Thursday, July 30, 2020

The CDC and APHL Next Generation Sequencing (NGS) Quality Initiative released the next set of ready-to-implement tools and documents. Adding to the collection of documents for Illumina Miseq, Oxford Nanopore’s MinION, and ThermoFisher’s Ion Torrent, this set of products includes SOPs for training personnel on Illumina’s MiniSeq and NextSeq. Additionally, this set provides tools and documents for two new Quality System Essentials (QSEs): Information Management and Assessment. Within this release are documents on best practices for documenting bioinformatic pipelines to support reproducibility under the Information Management QSE. This release also contains documents on identifying, developing, and applying key performance indicators to your sequencing assays and a Quality Management System Internal Assessment Tool to help laboratories assess compliance with quality system recommendations under the Assessment QSE.
Posted on Monday, March 9, 2020

A molecular biologist with CDC’s Division of Parasitic Diseases and Malaria (DPDM), Eldin Talundzic, PhD, recently returned from Peru, where he taught researchers to identify molecular markers associated with drug-resistant malaria using Advanced Molecular Detection (AMD) technologies. The training was part of CDC’s Malaria Resistance Surveillance project, or MaRS, which aims to develop and implement AMD tools in the U.S. and global partner laboratories to help identify and track drug-resistant malaria. The goal is to standardize and accelerate the flow of data used to track drug resistance, making it easier for decision makers to shape their malaria prevention and treatment guidance.
Posted on Thursday, January 30, 2020
The National Antimicrobial Resistance Monitoring System (NARMS), a public health surveillance system, tracks antibiotic-resistant bacteria that cause gastrointestinal and foodborne illness. Whole-genome sequencing (WGS), a new technology, has made this task easier. WGS lets scientists rapidly predict whether bacteria are resistant to antibiotics based on genetic information. In 2018, this information helped make the important distinction between two outbreaks occurring at the same time so public health officials could respond to each more effectively.
Posted on Wednesday, January 15th, 2020
To accompany a New England Journal of Medicine (NEJM) special report, Pathogen Genomics in Public Healthexternal icon, the Office of Advanced Molecular Detection (AMD) released a video, “Transforming Disease Detection,” which gives a brief introduction to next-generation sequencing (NGS) and explains how this technology can help solve outbreaks that, in the past, would not have been detectable.
