2019: Scientists use bacterial genome sequences to stop a deadly multi-facility outbreak
By Erin Scherer
The bacterial species Streptococcus pyogenes, or group A strep, is known for causing strep throat. It can also cause more serious invasive infections like “flesh-eating disease” or sepsis. Outbreaks of invasive group A strep infections may occur in long-term care facilities where older adults are at increased risk for infection. Outbreaks are caused by spreading the same bacterial strain from person-to-person. However, the extent to which individual outbreaks spread across multiple facilities remains unknown. Recent research in the Streptococcus Lab (StrepLab) at CDC is improving how scientists identify infections that are part of an outbreak. This new technology helped researchers detect and investigate a deadly multi-facility outbreak that might have otherwise been missed.
Using genome sequences to identify outbreaks
In the United States, a surveillance system including CDC, healthcare facilities, and labs tracks invasive group A strep infections. The Active Bacterial Core surveillance (ABCs) system collects clinical and demographic information about patients with bacterial infections. Participating ABCs sites then send patients’ bacterial strains to CDC’s StrepLab. Through start-up support from the Advanced Molecular Detection (AMD) program, StrepLab now sequences whole genomes of group A strep and other bacteria. Genome sequence information allows scientists to track new strains and antibiotic resistance trends.
StrepLab scientists realized they could also use genome sequences to find connections between invasive group A strep infections. Before genome sequencing, scientists only knew the type of group A strep strains, not if they were identical or near identical.1 The difference is like knowing whether a suspect and perpetrator have the same gender versus knowing they share the same DNA.
To find identical or near identical infections, StrepLab scientists use computers to:
- Compile thousands of group A strep genome sequences
- Organize them by type
- Find groups or “clusters” of highly similar genome sequences within each type
The resulting “cluster detection tool” accurately and efficiently identifies outbreaks of the same bacterial strain. Scientists have done this before for viruses, but viruses typically have much smaller genomes than bacteria.
Stopping an outbreak that might have gone undetected
While CDC’s scientists were developing the cluster detection tool in 2017, epidemiologists in Minnesota noticed an increase in invasive group A strep infections. The Minnesota Department of Health (MDH) found that the cases occurred across multiple long-term care facilities in the Minneapolis-St. Paul region. Could this be one outbreak involving multiple facilities?
The MDH Public Health Laboratory obtained potentially outbreak-related isolates, performed additional testing, and shared these isolates with StrepLab for sequencing and analysis. Running the genome sequences through the cluster detection tool at CDC showed the samples were near identical. StrepLab then analyzed older samples from ABCs and learned the problem was bigger than anyone suspected. Remarkably, the outbreak:
- Started the previous year in March 2016 and continued through October 2018
- Included more than 42 separate invasive group A strep infections
- Involved 19 long-term care facilities in Minnesota
- Resulted in nine deaths
What connected these infections? Epidemiologists in Minnesota reached out to more than 54 long-term care facilities to find clues. They identified multiple infection prevention lapses. In addition, facilities with outbreak infections were more likely to have shared a single wound care provider than facilities without outbreak infections.
While these findings are alarming, this investigation helped identify an invasive group A strep outbreak across multiple facilities and stop it. The true extent of the problem might never have been known or detected if not for:
- The ABCs system
- Capacity to sequence group A strep genomes supported by the AMD program
- StrepLab’s new cluster detection tool
StrepLab has since used the tool to detect multi-facility outbreaks of invasive group A strep infections in other states. This work demonstrates the potential for infections to spread between many facilities and that multi-facility outbreaks are more common than originally thought. These discoveries could help inform new policies to prevent deadly outbreaks.
Footnote:
1 Bacterial samples collected over the course of an outbreak might only be “near” identical because bacteria pick up genome mutations over time.
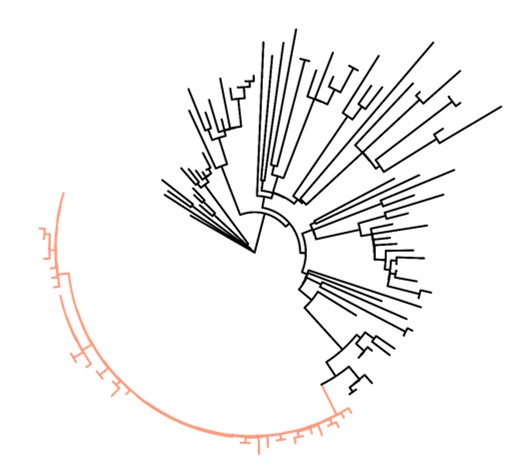
Group A strep strain sequences in Minnesota (March 2016 – August 2018)
This graph shows the similarities between different group A strep genome sequences. On this type of graph, more similar genome sequences cluster together on one “branch” with shorter outward stems. Sequences that are less similar appear on separate branches with longer outward stems. Each tip represents a different genome sequence.
Strains that were part of the multi-facility outbreak are near identical and cluster together on the orange branch. The stems are short indicating similarity. In contrast, other strains of the same type and time period are less similar to the outbreak strains and each other. They cluster on separate black branches with fewer sequences and longer stems.