2018 E. coli Outbreak Linked to Romaine Lettuce A
Posted June 28, 2018 3:30 PM EST
This outbreak appears to be over. E. coli is an important cause of illness in the United States. More information about E. coli, and steps people can take to reduce their risk of infection, can be found on the E. coli and Food Safety web page.
- This outbreak appears to be over as of June 28, 2018.
- CDC, public health and regulatory officials in several states, and the U.S. Food and Drug Administration (FDA) investigated a multistate outbreak of E. coli O157:H7 infections.
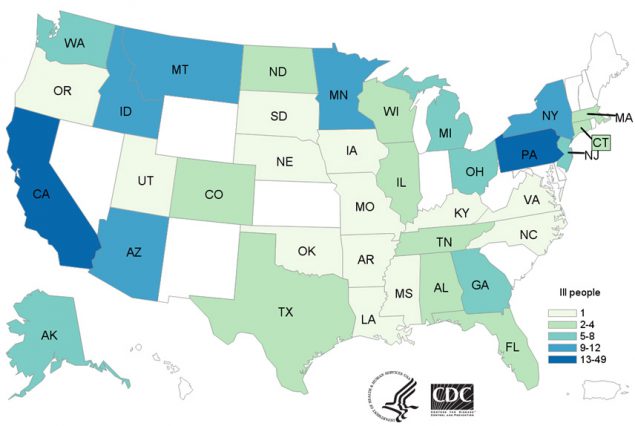
- 210 people infected with the outbreak strain were reported from 36 states.
- 96 people were hospitalized, including 27 people who developed a type of kidney failure called hemolytic uremic syndrome.
- 5 deaths were reported from Arkansas, California, Minnesota (2), and New York.
- Epidemiologic, laboratory, and traceback evidence indicated that romaine lettuce from the Yuma growing region was the likely source of this outbreak.
- CDC laboratory testing identified the outbreak strain of E. coli O157:H7 in canal water samples taken from the Yuma growing region. FDA is continuing to investigate the outbreak to learn more about how the E. coli bacteria could have entered the water and ways this water could have contaminated romaine lettuce.
- According to the FDA, the last shipments of romaine lettuce from the Yuma growing region were harvested on April 16, 2018, and the harvest season has ended. Contaminated lettuce that made people sick in this outbreak should no longer be available.
- The Public Health Agency of Canada (PHAC) identified ill people in several Canadian provinces infected with the same DNA fingerprint of E. coli O157:H7. On June 22, 2018, PHAC reported that the outbreak in Canada appears to be over.
- Consumers should follow these steps to help keep fruits and vegetables safer to eat.
- Read more on general ways to prevent E. coli infection. Important steps to take are to cook meat thoroughly, and wash hands after using the restroom or changing diapers, before and after preparing or eating food, and after contact with animals.
Introduction
CDC, public health and regulatory officials in several states, and the U.S. Food and Drug Administration (FDA) investigated a multistate outbreak of E. coli O157:H7 infections.
Public health investigators used the PulseNet system to identify illnesses that were part of this outbreak. PulseNet is the national subtyping network of public health and food regulatory agency laboratories coordinated by CDC. DNA fingerprinting is performed on E. coli bacteria isolated from ill people by using techniques called pulsed-field gel electrophoresis (PFGE) and whole genome sequencing (WGS). CDC PulseNet manages a national database of these DNA fingerprints to identify possible outbreaks. WGS gives a more detailed DNA fingerprint than PFGE. WGS performed on bacteria isolated from ill people in this outbreak showed that they were closely related genetically. This means that the ill people were more likely to share a common source of infection.
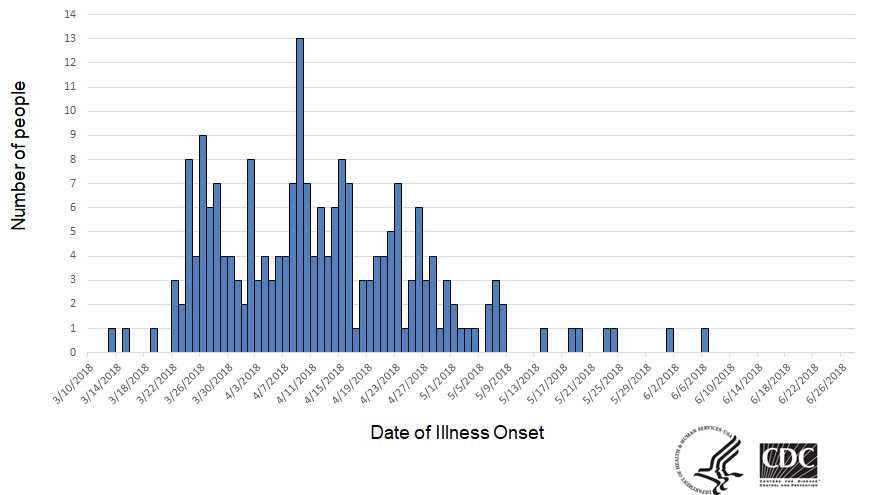
As of June 27, 2018, 210 people infected with the outbreak strain of E. coli O157:H7 were reported from 36 states. A list of the states and the number of cases in each can be found on the Case Count Map page. Illnesses started on dates ranging from March 13, 2018 to June 6, 2018. Ill people ranged in age from 1 to 88 years, with a median age of 28. Sixty-seven percent of ill people were female. Of 201 people with information available, 96 (48%) were hospitalized, including 27 people who developed hemolytic uremic syndrome, a type of kidney failure. Five deaths were reported from Arkansas, California, Minnesota (2), and New York.
WGS analysis of isolates from 184 ill people identified antibiotic resistance to chloramphenicol, streptomycin, sulfisoxazole, tetracycline, and trimethoprim-sulfamethoxazole. Standard antibiotic resistance testing of eight clinical isolates by CDC’s National Antimicrobial Resistance Monitoring System (NARMS) laboratory confirmed these findings. Isolates from four of those ill people also contained genes for resistance to ampicillin and ceftriaxone. These findings do not affect treatment guidance since antibiotics are not recommended for patients with E. coli O157 infections.
Investigation of the Outbreak
Epidemiologic, laboratory, and traceback evidence indicated that romaine lettuce from the Yuma growing region was the likely source of this outbreak.
In interviews, ill people answered questions about the foods they ate and other exposures they had before they became ill. Of the 166 people interviewed, 145 (87%) reported eating romaine lettuce in the week before their illness started. This percentage was significantly higher than results from a survey [PDF – 787 KB] of healthy people in which 46% reported eating romaine lettuce in the week before they were interviewed. Some people who became sick in this outbreak did not report eating romaine lettuce, but had close contact with someone else who got sick from eating romaine lettuce.
The FDA and state and local regulatory officials traced the romaine lettuce to many farms in the Yuma growing region. The FDA, along with CDC and state partners, started an environmental assessment in the Yuma growing region and collected samples of water, soil, and manure. CDC laboratory testing identified the outbreak strain of E. coli O157:H7 in water samples taken from a canal in the Yuma growing region. WGS showed that the E. coli O157:H7 found in the canal water is closely related genetically to the E. coli O157:H7 from ill people. Laboratory testing for other environmental samples is continuing. FDA is continuing to investigate to learn more about how the E. coli bacteria could have entered the water and ways this water could have contaminated romaine lettuce in the region.
According to the FDA, the last shipments of romaine lettuce from the Yuma growing region were harvested on April 16, 2018, and the harvest season there has ended. Contaminated lettuce that made people sick in this outbreak should no longer be available.
As of June 28, 2018, this outbreak appears to be over.